Annals of Cytology and Pathology
Specifics of determination of human biological age by blood samples using epigenetic markers
1The Institute of Genetics and Cytology National Academy of Sciences of Belarus, Minsk, Belarus
2Republican Scientific and Practical Centre «Cardiology», Minsk, Belarus
3Professor, State Institution «Republican Scientific and Practical Centre «Cardiology», Minsk, Belarus
Author and article information
Cite this as
Kipen V, Burakova A, Dobysh O, Zotova O, Bulgak A, et al. (2024) Specifics of determination of human biological age by blood samples using epigenetic markers. Ann Cytol Pathol. 2024; 9(1): 001-012. Available from: 10.17352/acp.000030
Copyright License
© 2024 Kipen V, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Our research focused on the selection of already known markers, as well as the search for other informative markers based on data made publicly available on the GEO NCBI platform (genome-wide DNA methylation projects using the Infinium Human Methylation 450K BeadChip (Illumina ©)).
The main objective of the study was to demonstrate that the accuracy of determining the biological age of a person in the presence of chronic diseases using linear-dependent methylation markers is comparable to the accuracy of determining the biological age of a healthy person.
Criminologists, as a rule, do not have information about the chronic diseases of a person who has left a biological trace at the scene (blood, for example). However, the lack of this information, as we have shown for some diseases, does not play a critical role in the precise determination of biological age.
Additionally, an obstacle was removed when transferring the information content of markers from Infinium Human Methylation 450K BeadChip chips to SNaPshot technology. The analysis was carried out on a sample of 236 Belarusians, for whom the methylation profile for 7 Cpg markers is presented. It is shown that the information content of markers is preserved
Our analysis shows the possibility of creating a universal test system for predicting biological age according to marker methylation. The system can be used in the work of most criminalists in the world with the same task.
Determination of biological age based on samples of biological fluids and tissue fragments plays an important role in forensic practice. It helps to limit the range of searches when identifying remains, to narrow the circle of suspects for saving time, as it is often a limiting factor in the investigation process. To determine the biological age of a person, the most sensitive, reproducible, and economically justified approach is based on detecting the level of DNA methylation in specific CpG dinucleotides [1-3].
Several methods have been proposed in the world for determining the biological age of an individual by the level of lifetime chemical modification of CpG-dinucleotides. These methods differ in the number of genetic markers under study and in the analysis method, with the claimed accuracy of 3-12 years [4-22]. Biological age, which reflects the degree of morphological and physiological development of the organism, in the context of DNA methylation, has a trend different from linear, but as close to it as possible. This is due to the hyper or hypofunctional expression of genes during the intensive growth of the body in the pre-pubertal and early pubertal periods, the presence of chronic diseases (bronchial asthma, multiple sclerosis, epilepsy, diabetes mellitus, cancer, etc.), normal gerontological processes, or the presence of alcoholic nicotine dependence and others [6,19,20,23,24]. Deviations in the change in the methylation profile from the linear trend for biological age associated with the growth and aging of the organism are most pronounced before 25 and after 60 years. The discrepancies between the biological and chronological age, which make it possible to assess the intensity of aging and the functional capabilities of an individual, are ambiguous in different phases of the development of the human body. In addition, the methylation level of specific CpG-dinucleotides may differ depending on the ethnogeographic origin of the individuals [25].
Modern methods for studying DNA methylation at the genome level suggest the use of one of two technological platforms for high-throughput analysis of nucleotide sequences - DNA hybridization on microarrays (microarray), or parallel clonal DNA sequencing (Massive parallel sequencing MPS or Next generation sequencing NGS). Illumina © hybridization microarrays remain the most popular platform for genomic DNA methylation analysis. Relatively low costs compared to whole genome sequencing positioned microarrays as a tool convenient for studying differentially methylated regions based on analysis of the methylation status of known CpG-sites in the human genome. For the Infinium HumanMethylation450 BeadChip (IHM 450K BeadChip), the largest array of primary data has been accumulated (in the form of the methylation level, expressed in % or fractions of a unit) for various types of biological samples (blood, individual blood cell fractions of, buccal epithelium, sperm, etc.), and for different ethnic groups or patients with a history of chronic diseases. The data is located in the Gene Expression Omnibus (GEO) Database repository (https://www.ncbi.nlm.nih.gov/geo/). Statistical analysis of raw data sets of the full genome DNA methylation profile will not only assess the accuracy of determining the biological age according to existing predictive models for independent samples differing in age, sex, or geography of residence of the studied groups within the framework of GEO projects but will also make it possible to identify previously characterized CpG-dinucleotides with high predictive potential.
The purpose of this work is to assess the evaluation of ethnoregional, sex, and other factors in the context of determining biological age from blood samples using methylation data of CpG-dinucleotides. It is based on the analysis of the primary data of the whole genome DNA methylation profile from GEO DataSets NCBI, as well as to check the revealed patterns in the contribution of highly informative CpG-dinucleotides in the accuracy of determining the biological age of individuals from the Republic of Belarus.
Materials and methods
DNA samples in silico
Information on the DNA methylation level for blood samples is available on the NCBI GEO datasets platform for 8 projects: GSE40279, GSE42861, GSE51032, GSE50660, GSE55763, GSE77696, GSE106648, GSE125105. The main criterion for selecting projects is the availability of information on the DNA methylation profile for at least 250 people. After a two-stage mathematical preparation of the primary data, the number of healthy individuals of various ethnographic origins was 4251 people, with a history of acute or chronic diseases of 1685 people. The biological age range was from 17 to 93 years. The number of blood samples from men is 3169, and from women 2766, for 3 samples there was no information on sex.
DNA samples of individuals from the Republic of Belarus
Blood samples from 236 individuals aged 18 to 93 years were obtained after signing an informed consent approved by the Bioethics Committee of the Institute of Genetics and Cytology of the National Academy of Sciences of Belarus (Protocol No.8, 2017). BD Vacutainer K2E tubes were used to collect venous blood. DNA was isolated using MagMAX ™ DNA Multi-Sample Kit (ThermoFisher, USA) according to the manufacturer’s recommendations. These purification kits use MagMAX™ magnetic bead-based nucleic acid isolation technology to produce high yields of purified DNA, free from inhibitors that may affect downstream PCR. The quality and quantity of DNA was analyzed using a NanoPhotometer® N50 spectrophotometer (IMPLEN, USA).
CpG-dinucleotides for modeling
We analyzed in silico data for 16 CpG-dinucleotides. The predictive potential of 10 CpG-dinucleotides was confirmed in a study [17]: cg02872426 (DDO gene), cg06784991 (ZYG11A gene), cg06874016 (NKIRAS2 gene), cg07553761 (TRIM59 gene), cg11807280 (MEIS1-AS3 gene), cg14361627 (KLF14 gene), cg16054275 (F5 gene), cg16867657 (ELOVL2 gene) cg18473521 (HOXC4 gene), cg25410668 (RPA2 gene).
We independently determined a high prognostic potential in silico based on bioinformatics analysis of data from GEO projects for 6 CpG-dinucleotides: cg05213896 (IL4I1 gene), cg08128734 (RASSF5 gene), cg08468401, cg19283806 (CCDC102B gene), cg2245H269 (FHL2 gene), cg24079702 (FHL2 gene). Information on the methylation level of CpG-dinucleotides and the characteristics of individuals included in the analysis in silico is presented in “Supplementary materials.docx / Sheet 1”.
Determination of methylation level using SNaPshot
Analysis of the methylation level for CpG-dinucleotides was performed using SNaPshot technology (Applied Biosystems ™, USA). Primers and SBE-oligonucleotides (Single-base extension SBE) for CpG-dinucleotides are presented in Table 1. Primers for amplification of bisulfite-converted genomic DNA were developed using the BiSearch program (http://bisearch.enzim.hu/).
PCR was performed in a volume of 20 μl, containing 10-15 ng of bisulfite-converted genomic DNA, 1U ArtStart DNA polymerase (ArtBioTeсh, Belarus), 2.0 μl of 10x PCR buffer (containing Mg2 + at a concentration of 2.0 mM), 0.08 mM each deoxynucleotide (dATP, dGTP, dCTP, dTTP), 0.4-1.0 μM R- and F-primer. Bisulfite-converted genomic DNA was obtained by modifying 200-500 ng of genomic DNA using the MethylEdge® kit (Promega, USA). PCR was performed in an Applied Biosystems ProFlex PCR System thermal cycler (Thermo Fisher Scientific, USA): 95 °С - 4 min; (94 °С - 20 s, 56 °С –30 s, 72 °С - 45 s) - 34 cycles; 72 °C –7 min. Then 5 μl of each PCR product was purified using the Exo-CIP ™ Rapid PCR Cleanup Kit (NEB, USA).
SBE was performed using 3 µL of the purified PCR product, 0.2–0.4 mM SBE oligonucleotide, and an SNaPshot kit (Applied Biosystems, USA). Then 10 μl of each SBE product was purified using 1 μl FastAP Thermosensitive Alkaline Phosphatase (ThermoFisher, USA). SBE products were analyzed using an ABI PRISM 3500 genetic analyzer and GeneMapper® 5.0 software (Applied Biosystems, USA). The percentage methylation value (0-100%) for each CpG-dinucleotide was calculated by dividing the fluorescence intensity value for C/G nucleotides (detection of unconverted methylated DNA) by the fluorescence intensity value for C/G nucleotides plus T/A (detection of converted unmethylated DNA).
Statistical data analysis
The first stage in preparing GEO project data for mathematical analysis is excluding values outside the range calculated by the formula:
[(X25 – 1,5 * (X75 – X25), (X75 + 1,5 * (X75 – X25))]
This range is calculated separately for each GEO project.
The second stage is the normalization of the data remaining after the first stage using a nonlinear transformation within [-1, 1] by the formula:
(X- Median)/SQRT (SUMM ((Х- Median) ^ 2))
The second stage is performed for the data array obtained in the first stage. Thus, the two-stage data preparation made it possible to minimize the contribution of extreme values as much as possible.
We used the same data preparation scheme for statistical analysis to establish the DNA methylation level values from 16 CpG-dinucleotides of blood samples from Belarusian individuals.
Using the SPSS v.20.0 program (IBM, USA), we calculated rank correlation coefficients (R) via the bootstrap function for 1000 samples (with bias correction and acceleration) and calculated a 95% confidence interval. Also were corrected values of the coefficients of determination (R^2), equal to the proportion of the variance of the dependent variable “biological age” due to the influence of independent variables (the level of methylation of CpG-dinucleotides); Mean Absolute Deviation (MAD) and root mean square errors (RMS Error, RMSE) for regression models.
Results and discussions
As a rule, projects to assess the genome-wide methylation profile using the IHM 450K BeadChip target a cohort of people who represent a specific cross-section of the population of a particular region or ethnicity. Researchers aim to find relations between the DNA methylation profile and disease as applied to a specific country or geographic region. We carried out comparative studies and characterized the correlation coefficients for the 16 CpG-dinucleotides listed above, depending on the ecoregional and sex identity of individuals, as well as on the presence of chronic diseases in history (rheumatoid arthritis, HIV, multiple sclerosis, depressive disorders, oncological diseases) or bad habits (nicotine addiction).
Correlation coefficients of DNA methylation with biological age depending on the ethnogeographic factor
Correlation coefficients (R) for 16 CpG-dinucleotides were calculated within 8 GEO projects within the countries of the European (UK, Italy, Sweden, Germany) and North American (USA) regions are presented in Table 2. The information is used only for healthy individuals, taking into account ethno geographic status and without regard to sex. The number of persons for the European region was 3579 (Great Britain – 2614, Italy – 362, Sweden – 430, Germany – 173), and for the North American region – 672.
For three CpG-dinucleotides, the R-values were the most reproducible, as evidenced by the low values of the standard deviation – cg19283806 (-0.571 ± 0.068), cg25410668 (0.492 ± 0.069) and cg16867657 (0.810 ± 0.073), while for two of them – cg19283806 and cg16867657 shows the largest absolute values of R. The largest fluctuation of R-values is shown for the CpG-dinucleotides cg18473521 (standard deviation – 0.151), cg11807280 (0.146) and cg24079702 (0.135).
Correlation coefficients of DNA methylation with biological age depending on sex
The R coefficients for 16 CpG-dinucleotides were calculated within 6 GEO projects and are presented in Table 3. The number of males (sample “M”) was 2247 individuals, female (sample “F”) – 1777 individuals. The most reproducible R-values for males are shown for cg25410668 (0.448 ± 0.082), cg08128734 (-0.453 ± [0.091]), cg16867657 (0.749 ± 0.095), cg16054275 (-0.462 ± [0.098]) and cg08468401 (-0.399 ± [ 0.099]); for females - for cg19283806 (-0.565 ± [0.066]), cg25410668 (0.519 ± 0.077), cg02872426 (-0.374 ± [0.077]), cg16867657 (0.819 ± 0.081) and cg16054275 (-0.458 ± [0.091]).
Differences between R - values depending on sex ranged from 0.002 to 0.071. The smallest fluctuation in R-values is shown for CpG-dinucleotides cg22454769 (difference – 0.002), cg16054275 (0.004), cg18473521 (0.011), cg14361627 (0.014), and cg19283806 (0.016).
Calculation of determination coefficients (R^2), MAD, and RSME for regression models of predicting biological age
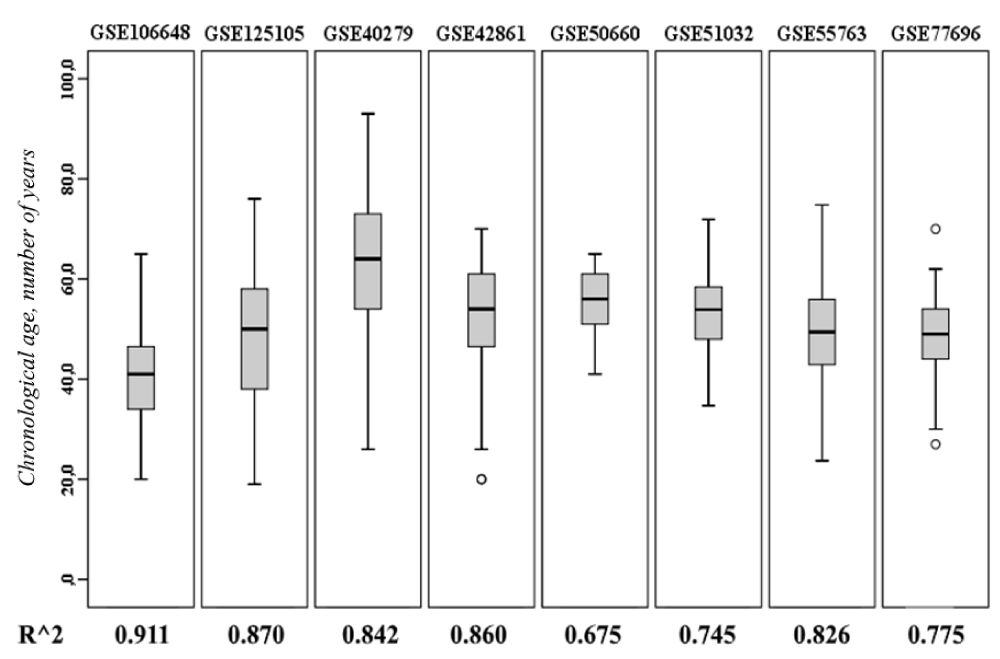
Based on the data on the methylation level of 16 CpG-dinucleotides, we adjusted determination coefficients for multiple linear regression for GEO projects (according to Table 1). According to the data presented in (Figure 1), it can be seen that the narrower the range for the indicator “Chronological age, number of years” appeared in the study (for example, for projects GSE51032 or GSE50660), the less the adjusted R^2 was. As known, the regression model is able to adequately (with the calculated level of accuracy) predict the dependent variable (biological age) when modeling only in the analyzed range of values; therefore, expanding the scope for the dependent variable is able to stabilize the model.
Thus, the adjusted R^2 values were in the range 0.675-0.911, and for GEO projects with the widest age range – GSE125105, GSE40279, and GSE55763 - the percentage of explained variation for the dependent variable was at least 82.6%.
According to 8 GEO projects, CpG-dinucleotides had a different effect on the change in the coefficient of determination R^2 (Table 4). The largest contribution to the percentage of explained variance of the dependent variable in the regression model equation belonged to CpG-dinucleotides: cg16867657 - mean value R^2 = 0.669, cg14361627– 0.056 and cg19283806 - 0.044. The predictive potential for the CpG dinucleotide cg19283806 proved comparable to the value for cg14361627. The high predictive potential of this CpG dinucleotide was also shown in the study [26].
When models for predicting biological age were created, we used an approach according to which the dependence of the level of DNA methylation on the age of individuals was considered linear. In our view, its use is justified under the condition of a relatively large number of individuals in the study when analyzing contrasting age samples.
We found that the percentage of the explained variance R^2 when modeling multiple linear regression using the stepwise selection function (inclusion with a probability F < 0.05, an exclusion with a probability F > 0.10) varied in the range 0.676-0.911. MAD values were in the range of 1.92-3.26 years (Table 4). Each model for predicting biological age included a different number of CpG-dinucleotides - from 5 for GEO77696 to 10 for GEO55763.
It is known that the R^2, MAD, and RMSE indices reflect the overall accuracy of the model and make it possible to compare the models with each other, but they poorly characterize the predictive accuracy of the dependent variable (biological age) for a particular sample. In (Figure 2) provides information on the number of individuals, expressed as a percentage (%) within each GEO project, for which the predicted values of biological age were calculated using the regression model (Table 4) within a given error - “≤ 2 years”, “> 2 and ≤ 4 years ”,“> 4 and ≤ 6 years ”,“> 6 and ≤ 8 years ”,“> 8 and ≤ 10 years ”and“> 10 years ”.
Thus, the percentage of predicted biological age values with an error of ≤ 4 years ranged from 58.6% (for the GEO project GSE55763) to 80.3% (for the GEO project GSE125105), with an error of ≤ 6 years – 76.8-96.1%. The number of cases with an error in predicting the biological age of more than 8 years on average for eight GEO projects was less than 5.0% (Figure 2).
As can be seen from (Figure 3), in three age groups “≤ 40 years old”, “> 40 and ≤ 60 years old”, and “> 60 years old” the percentage of predicted values of biological age with an error of ± 6 years was 81.9 ± 12.2%, 90.6 ± 5.6%, and 83.9 ± 10.7%, respectively. The highest percentage of correct calculations (± 6 years) falls in the age range “> 40 and ≤ 60 years.” In the sample “> 60 years old,” the error in predicting biological age gradually increases. This may be due to an increase in the variance for the level of methylation of the analyzed CpG sites with age during aging, which is due to a wide range of reactions of the human body in normal and pathological gerontological processes.
Calculation of the coefficients of determination (R^2), MAD and RSME for regression models for predicting biological age, depending on the anamnesis
The question of the influence of pathological processes in the body on changes in the methylation level of the analyzed CpG-dinucleotides in determining the biological age of an individual is important. To develop a method for determining the age of an unknown individual, which can be used in forensic practice, it is necessary to use those CpG-dinucleotides, the methylation level of which does not critically differ in healthy and sick individuals. The key characteristic of the CpG dinucleotide for assessing its predictive potential in determining biological age is the value of the determination coefficient R, the differences of which in the group of sick and healthy individuals must be identified.
In this regard, we analyzed information from open sources regarding the level of DNA methylation for 16 CpG-dinucleotides for pathological conditions: rheumatoid arthritis (GSE42861, n = 306, age range 22.0-69.0 years); HIV (GSE77696, n = 229, 25.0-70.0 years); multiple sclerosis (GSE106648, n = 130, 18.0-66.0 years); depressive disorders (GSE125105, n = 420, 17.0-87.0 years); oncological diseases (GSE51032: breast cancer, n = 191; colorectal cancer, n = 68; other primary tumors, n = 101; 35.0-72.0 years), as well as for individuals with nicotine addiction (quit smoking after prolonged period - GSE50660, n = 221; continuing smoking - GSE50660, n = 19; 44.0-65.0 years).
In (Figure 4) provides information on regression models for predicting biological age and their characteristics for the indicated pathological conditions.
The calculated MAD values for the studied pathological conditions were arranged in decreasing order in the following sequence: HIV - 3.9 years, depressive disorders - 3.3 years, rheumatoid arthritis - 2.7 years, oncological diseases - 2.5 years, multiple sclerosis - 1.9 years old. For individuals with nicotine addiction, the accuracy of predicting biological age was 3 years.
Only for patients with HIV, the MAD values were 3.9 years, and the difference between sick and healthy individuals was more than one year. For other pathological conditions, the difference in MAD values between healthy individuals and patients was less than one year.
Thus, pathological conditions do not have a critical impact on determining the biological age of a person by the methylation level of the studied CpG-dinucleotides.
Calculation of the probability of attributing an unknown sample to a specific age group based on DNA methylation data
Often, for forensic practice, when determining the estimated age of an unknown individual, the question is not about a specific age, but about the assignment of a given subject to a certain age group: “under 20” or “over 20”, “under 30” or “over 30” etc. In this case, the accuracy of assigning an unknown individual to a specific group based on the results of DNA methylation analysis will be higher than when answering the question about the true value of the biological age. At the same time, to clarify the predicted age, a two-stage scheme can be used: 1) assigning an unknown sample to a certain age group (with a level of accuracy acceptable for specific tasks of forensic science); 2) predicting the value of biological age in years (with a level of accuracy within the predictive model) already within the age group.
Therefore, depending on the type of division of samples array by age categories, the accuracy of assigning a particular sample varies in a wide range (Figure 5). With a probability of 99.21 ± 0.86%, it can be concluded that the age of the unknown resident, established using 5-10 СpG dinucleotides, is more than 30 years, with a probability of 97.61 ± 1.74%, it is more than 40 years, 91.56 ± 5.19% - more than 50 years, etc. The average classification accuracy within each boundary age point “30” - “60” was 87.05 ± 3.82%.
Thus, the conducted bioinformatics and statistical analysis of GEO projects allows us to draw a number of conclusions. First, of the 16 analyzed CpG-dinucleotides, cg16867657, cg14361627, and cg19283806 have the highest predictive potential. Secondly, for all eight regression models within the GEO projects, comparable accuracy in predicting biological age was shown based on the values of MAD (1.92-3.26) and RMSE (1.94-3.29). At the same time, all 3 CpG-dinucleotides with the highest predictive potential are involved in the models for seven of the eight GEO projects. Thirdly, concomitant factors (sex, ethnogeographic affiliation, the presence of pathological conditions) do not significantly affect the accuracy of predicting biological age when using the analyzed CpG-dinucleotides.
However, it should be noted that the results obtained have a number of limitations on interpretation and extrapolation. Thus, it is known that the results obtained using the IHM 450K BeadChip technology (Illumina, USA) may not coincide with the results obtained using the SNaPshot technology (Applied Biosystems, USA), and, thus, CpG-dinucleotides determined on the basis of bioinformatic analysis as highly informative (in R > 0.5) may not show themselves when studying specific groups using the SNaPshot microsequencing technology. In this regard, for individuals from the Republic of Belarus, we determined the methylation levels of 7 CpG-dinucleotides. The predictive potential of which according to the results of the analysis (Table 4) was maximum: cg07553761, cg14361627, cg16054275, cg16867657, cg19283806, cg24079702, and cg25410668.
In general, our data on the level of DNA methylation for 7 CpG-dinucleotides for the Belarus sample are comparable to those for the largest GEO project, GSE55769, despite the statistically significant differences (Figure 6). According to the value of the correlation coefficients R with biological age, CpG-dinucleotides were arranged in the following sequence (in decreasing order of the absolute value of R): cg19283806 (R = -0.739, p = 5.57E-42), cg16867657 (0.687, 2.37E-34), cg07553761 (0.654, 3.87E-30), cg14361627 (0.642, 8.25E-29), cg25410668 (0.559, 8.34E-21), cg16054275 (-0.378, 2.02E-09) and cg24079702 (0.170, 8,95E-03).
By analogy with the previous analysis, statistical data preprocessing was carried out and the regression model was calculated, which is graphically presented in (Figure 7). The largest contribution to the variance of the variable “Biological age” is made by the CpG dinucleotide cg19283806 (gene CCDC102B) - no less than 62.9%. Next are CpG-dinucleotides in the order of decreasing influence on the variable “Biological age” in the regression model: cg14361627 (KLF14 gene) – + 13.3%, cg16867657 (EVOLV2 gene) – + 6.1%, cg07553761 (TRIM59 gene) – + 1.0%, cg25410668 (PRA2 gene) – + 0.7%, cg24079702 (FHL2 gene) – +0.7, cg16054275 (F5 gene) – + 0.5%.
Our proposed model for predicting age based on the methylation profile CpG-dinucleotides of blood is relatively simple, since a small number of markers are used in the analysis and the technique developed using them can be used in forensic laboratories of a molecular genetic orientation. The age prediction error for the model we calculated corresponds to similar studies [27-36].
Conclusion
Based on the data presented in the public domain on the GEO NCBI platform for 8 projects to determine the full genome DNA methylation profile using the Infinium Human Methylation 450K BeadChip (Illumina ©) - GSE40279, GSE42861, GSE51032, GSE50660, GSE55763, GSE77696, GSE1051048 with a total number of individuals of more than 4 thousand (without a history of chronic and acute diseases), we calculated the correlation coefficients (R) with biological age for 16 CpG-dinucleotides. Also, we calculated the corrected coefficients of determination (R^2), MAD, and RMSE for comparisons and characteristics of multivariate linear regression equations.
Based on bioinformatics and statistical analysis, we have shown that for individuals without a history of chronic or acute diseases, regardless of ethnic geographic and sexual factors, CpG-dinucleotides cg14361627 (gene KLF14), cg16867657 (gene ELOVL2) and cg19283806 (gene CCDC102B), on average they are able to explain the variance of the variable “Biological age” by 35.6 ± 10.4%, 65.0 ± 11.8%, and 33.0 ± 8.7%, respectively. For individuals from the Republic of Belarus, for the CpG dinucleotide cg19283806 (CCDC102B), the percentage of the explained variance of the variable “Biological age” turned out to be the maximum - 62.7%, the share of cg14361627 (KLF14 gene) and cg16867657 (ELOVL2 gene) accounted for + 13.3% and + 6.1%, respectively. In total, these three CpG-dinucleotides can explain at least 80% of the variation in the biological age of a person.
The methodology for determining biological age by establishing a DNA methylation profile based on a limited number of CpG-dinucleotides (5-10 pcs). The prognostic potential which has been confirmed in a number of studies and demonstrated by us on samples of Belarussian individuals, is universal. It is possible to provide sufficiently accurate information about the estimated age of an individual or about belonging to a particular age group, regardless of the ethno-geographic status of an unknown person, sex, or the presence of a number of chronic diseases.
Financing. The study was carried out within the framework of the Scientific and Technical Program of the Union State “Development of innovative geno geographic and genomic technologies for identification of personality and individual characteristics of a person based on the study of gene pools of the regions of the Union State” (“DNA-identification”) in the context of Activity No. 2 “Development of a method for determining the probable age of an individual according to the characteristics of his DNA “(2017-2021).
All procedures performed in human research comply with the ethical standards of the institutional and/or national committee on research ethics and the Declaration of Helsinki (1964) and its subsequent amendments or comparable standards of ethics. Voluntary informed consent was obtained from each of the participants included in the study.
- Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y, Deconde R, Chen M, Rajapakse I, Friend S, Ideker T, Zhang K. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013 Jan 24;49(2):359-367. doi: 10.1016/j.molcel.2012.10.016. Epub 2012 Nov 21. PMID: 23177740; PMCID: PMC3780611.
- Kurdyukov S, Bullock M. DNA Methylation Analysis: Choosing the Right Method. Biology (Basel). 2016 Jan 6;5(1):3. doi: 10.3390/biology5010003. PMID: 26751487; PMCID: PMC4810160.
- Goel N, Karir P, Garg VK. Role of DNA methylation in human age prediction. Mech Ageing Dev. 2017 Sep;166:33-41. doi: 10.1016/j.mad.2017.08.012. Epub 2017 Aug 24. PMID: 28844970.
- Koch CM, Wagner W. Epigenetic-aging-signature to determine age in different tissues. Aging (Albany NY). 2011 Oct;3(10):1018-27. doi: 10.18632/aging.100395. PMID: 22067257; PMCID: PMC3229965.
- Bocklandt S, Lin W, Sehl ME, Sánchez FJ, Sinsheimer JS, Horvath S, Vilain E. Epigenetic predictor of age. PLoS One. 2011;6(6):e14821. doi: 10.1371/journal.pone.0014821. Epub 2011 Jun 22. PMID: 21731603; PMCID: PMC3120753.
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115. Erratum in: Genome Biol. 2015;16:96. PMID: 24138928; PMCID: PMC4015143.
- Weidner CI, Lin Q, Koch CM, Eisele L, Beier F, Ziegler P, Bauerschlag DO, Jöckel KH, Erbel R, Mühleisen TW, Zenke M, Brümmendorf TH, Wagner W. Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol. 2014 Feb 3;15(2):R24. doi: 10.1186/gb-2014-15-2-r24. PMID: 24490752; PMCID: PMC4053864.
- Lee HY, Lee SD, Shin KJ. Forensic DNA methylation profiling from evidence material for investigative leads. BMB Rep. 2016 Jul;49(7):359-69. doi: 10.5483/bmbrep.2016.49.7.070. PMID: 27099236; PMCID: PMC5032003.
- Bekaert B, Kamalandua A, Zapico SC, Van de Voorde W, Decorte R. Improved age determination of blood and teeth samples using a selected set of DNA methylation markers. Epigenetics. 2015;10(10):922-30. doi: 10.1080/15592294.2015.1080413. Epub 2015 Aug 17. PMID: 26280308; PMCID: PMC4844214.
- Zbieć-Piekarska R, Spólnicka M, Kupiec T, Makowska Ż, Spas A, Parys-Proszek A, Kucharczyk K, Płoski R, Branicki W. Examination of DNA methylation status of the ELOVL2 marker may be useful for human age prediction in forensic science. Forensic Sci Int Genet. 2015 Jan;14:161-7. doi: 10.1016/j.fsigen.2014.10.002. Epub 2014 Oct 14. PMID: 25450787.
- Zbieć-Piekarska R, Spólnicka M, Kupiec T, Parys-Proszek A, Makowska Ż, Pałeczka A, Kucharczyk K, Płoski R, Branicki W. Development of a forensically useful age prediction method based on DNA methylation analysis. Forensic Sci Int Genet. 2015 Jul;17:173-179. doi: 10.1016/j.fsigen.2015.05.001. Epub 2015 May 5. PMID: 26026729.
- Park JL, Kim JH, Seo E, Bae DH, Kim SY, Lee HC, Woo KM, Kim YS. Identification and evaluation of age-correlated DNA methylation markers for forensic use. Forensic Sci Int Genet. 2016 Jul;23:64-70. doi: 10.1016/j.fsigen.2016.03.005. Epub 2016 Mar 17. PMID: 27017110.
- Zubakov D, Liu F, Kokmeijer I, Choi Y, van Meurs JBJ, van IJcken WFJ, Uitterlinden AG, Hofman A, Broer L, van Duijn CM, Lewin J, Kayser M. Human age estimation from blood using mRNA, DNA methylation, DNA rearrangement, and telomere length. Forensic Sci Int Genet. 2016 Sep;24:33-43. doi: 10.1016/j.fsigen.2016.05.014. Epub 2016 May 24. PMID: 27288716.
- Alghanim H, Antunes J, Silva DSBS, Alho CS, Balamurugan K, McCord B. Detection and evaluation of DNA methylation markers found at SCGN and KLF14 loci to estimate human age. Forensic Sci Int Genet. 2017 Nov;31:81-88. doi: 10.1016/j.fsigen.2017.07.011. Epub 2017 Aug 7. PMID: 28854399.
- Alsaleh H., McCallum N.A., Halligan D.L., Haddrill P.R. A multi-tissue age prediction model based on DNA methylation analysis / Forensic Science International: Genetics Supplement Series. 2017. 6: 62-64. DOI: 10.1016/j.fsigss.2017.09.056
- Mawlood SK. Pickard B.S. Methylation Status and Human Age at Tree Autosomal Loci: A New Forensic Profling Tool / Journal of Forensic and Crime Studies. 2017; e1-e8.
- Naue J, Hoefsloot HCJ, Mook ORF, Rijlaarsdam-Hoekstra L, van der Zwalm MCH, Henneman P, Kloosterman AD, Verschure PJ. Chronological age prediction based on DNA methylation: Massive parallel sequencing and random forest regression. Forensic Sci Int Genet. 2017 Nov;31:19-28. doi: 10.1016/j.fsigen.2017.07.015. Epub 2017 Aug 1. PMID: 28841467.
- Thong Z, Shun Chan XL, Tan JYY, Loo ES, Choong Syn CK. Evaluation of DNA methylation-based age prediction on blood / Forensic Science International: Genetics Supplement Series. 2017; 6: e249-e251. DOI: 10.1016/j.fsigss.2017.09.095
- Vidaki A, Kayser M. From forensic epigenetics to forensic epigenomics: broadening DNA investigative intelligence. Genome Biol. 2017; 21: e1-e13. DOI: 10.1186/s13059-017-1373-1.
- Freire-Aradas A, Phillips C, Girón-Santamaría L, Mosquera-Miguel A, Gómez-Tato A, Casares de Cal MÁ, Álvarez-Dios J, Lareu MV. Tracking age-correlated DNA methylation markers in the young. Forensic Sci Int Genet. 2018 Sep;36:50-59. doi: 10.1016/j.fsigen.2018.06.011. Epub 2018 Jun 13. PMID: 29933125.
- Naue J, Sänger T, Hoefsloot HCJ, Lutz-Bonengel S, Kloosterman AD, Verschure PJ. Proof of concept study of age-dependent DNA methylation markers across different tissues by massive parallel sequencing. Forensic Sci Int Genet. 2018 Sep;36:152-159. doi: 10.1016/j.fsigen.2018.07.007. Epub 2018 Jul 7. PMID: 30031222.
- Smeers I, Decorte R, Van de Voorde W, Bekaert B. Evaluation of three statistical prediction models for forensic age prediction based on DNA methylation. Forensic Sci Int Genet. 2018 May;34:128-133. doi: 10.1016/j.fsigen.2018.02.008. Epub 2018 Feb 9. PMID: 29477092.
- Rana AK. Crime investigation through DNA methylation analysis: methods and applications in forensics / Egyptian Journal of Forensic Sciences. 2018; 8: e1-e17. DOI 10.1186/s41935-018-0042-1
- Vidaki A, Ballard D, Aliferi A, Miller TH, Barron LP, Syndercombe Court D. DNA methylation-based forensic age prediction using artificial neural networks and next generation sequencing. Forensic Sci Int Genet. 2017 May;28:225-236. doi: 10.1016/j.fsigen.2017.02.009. Epub 2017 Feb 28. PMID: 28254385; PMCID: PMC5392537.
- Fleckhaus J, Freire-Aradas A, Rothschild MA, Schneider PM. Impact of genetic ancestry on chronological age prediction using DNA methylation analysis / Forensic Science International: Genetics. Supplement Series. 2017; 6: e399-e400. DOI: 10.1016/j.fsigss.2017.09.162
- Pan C, Yi S, Xiao C, Huang Y, Chen X, Huang D. The evaluation of seven age-related CpGs for forensic purpose in blood from Chinese Han population. Forensic Sci Int Genet. 2020 May;46:102251. doi: 10.1016/j.fsigen.2020.102251. Epub 2020 Jan 24. PMID: 32006895.
- Aliferi A, Ballard D, Gallidabino MD, Thurtle H, Barron L, Syndercombe Court D. DNA methylation-based age prediction using massively parallel sequencing data and multiple machine learning models. Forensic Sci Int Genet. 2018 Nov;37:215-226. doi: 10.1016/j.fsigen.2018.09.003. Epub 2018 Sep 8. PMID: 30243148.
- Alsaleh H, Haddrill PR. Identifying blood-specific age-related DNA methylation markers on the Illumina MethylationEPIC® BeadChip. Forensic Sci Int. 2019 Oct;303:109944. doi: 10.1016/j.forsciint.2019.109944. Epub 2019 Sep 12. PMID: 31546163.
- Anaya Y, Yew P, Roberts KA, Hardy WR. DNA methylation of decedent blood samples to estimate the chronological age of human remains. Int J Legal Med. 2021 Nov;135(6):2163-2173. doi: 10.1007/s00414-021-02650-8. Epub 2021 Jul 10. PMID: 34245337.
- Daunay A, Baudrin LG, Deleuze JF, How-Kit A. Amp; How-Kit, AEvaluation of Six Blood-Based Age Prediction Models Using DNA Methylation Analysis by Pyrosequencing. Sci. Rep. 2019. 9(1): 8862. DOI:10.1038/s41598-019-45197-w
- Dias HC, Cordeiro C, Pereira J, Pinto C, Real FC, Cunha E, Manco L. DNA methylation age estimation in blood samples of living and deceased individuals using a multiplex SNaPshot assay. Forensic Sci Int. 2020 Jun;311:110267. doi: 10.1016/j.forsciint.2020.110267. Epub 2020 Apr 16. PMID: 32325350.
- Gao X, Liu S, Song H, Feng X, Duan M, Huang L, Zhou F. AgeGuess, a Methylomic Prediction Model for Human Ages. Front Bioeng Biotechnol. 2020 Mar 10;8:80. doi: 10.3389/fbioe.2020.00080. PMID: 32211384; PMCID: PMC7075810.
- Jung SE, Lim SM, Hong SR, Lee EH, Shin KJ, Lee HY. DNA methylation of the ELOVL2, FHL2, KLF14, C1orf132/MIR29B2C, and TRIM59 genes for age prediction from blood, saliva, and buccal swab samples. Forensic Sci Int Genet. 2019 Jan;38:1-8. doi: 10.1016/j.fsigen.2018.09.010. Epub 2018 Sep 29. PMID: 30300865.
- Ryan J, Wrigglesworth J, Loong J, Fransquet PD, Woods RL. A Systematic Review and Meta-analysis of Environmental, Lifestyle, and Health Factors Associated With DNA Methylation Age. J Gerontol A Biol Sci Med Sci. 2020 Feb 14;75(3):481-494. doi: 10.1093/gerona/glz099. PMID: 31001624; PMCID: PMC7328212.
- Woźniak A, Heidegger A, Piniewska-Róg D, Pośpiech E, Xavier C, Pisarek A, Kartasińska E, Boroń M, Freire-Aradas A, Wojtas M, de la Puente M, Niederstätter H, Płoski R, Spólnicka M, Kayser M, Phillips C, Parson W, Branicki W; VISAGE Consortium. Development of the VISAGE enhanced tool and statistical models for epigenetic age estimation in blood, buccal cells and bones. Aging (Albany NY). 2021 Mar 11;13(5):6459-6484. doi: 10.18632/aging.202783. Epub 2021 Mar 11. PMID: 33707346; PMCID: PMC7993733.
- Xiao C, Yi S, Huang D. Genome-wide identification of age-related CpG sites for age estimation from blood DNA of Han Chinese individuals. Electrophoresis. 2021 Aug;42(14-15):1488-1496. doi: 10.1002/elps.202000367. Epub 2021 Jun 10. PMID: 33978960.
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


.gif)
.gif)





 Save to Mendeley
Save to Mendeley
