Annals of Molecular and Genetic Medicine
Expression of blaCTX-M2and invA genes of Salmonella Heidelberg isolated from poultry by Qpcr
Gabriella Bassi das Neves1, Denise Nunes Araújo1,2, Eduarda Pick2, Dinael Simão Bitner2, Maiara Cristiane Brisola1, Regiane Boaretto Crecencio1, and Lenita Moura Stefani1*
Cite this as
Stefani LM (2020) Expression of blaCTX-M2and invA genes of Salmonella Heidelberg isolated from poultry by Qpcr. Ann Mol Genet Med 4(1): 006-011. DOI: 10.17352/amgm.000006Salmonellosis is a disease caused by a bacterium Salmonella, a gram negative bacilli found in many environments, responsible for significant economic losses in poultry, and of great impact on public health. Among more than 2500 serovars, S. Heidelberg seems to be more invAsive causing disease of greater severity than other serovars. The objective of this study was to investigate, through real-time PCR (qPCR), differences in the expression of a virulent gene (invA) and an antibiotic resistance gene (blaCTXM-2) of S. Heidelberg isolated from poultry meat (slaughterhouses) and drag swabs (field). Even though all isolates showed the presence of the invA gene, there were differences in the expression among the isolates, where isolates from the field showed greater expression of invA compared to samples isolated from meat products. On the other hand, isolates from the slaughterhouses showed greater expression of the blaCTX-M2than those isolated from field samples.
Introduction
Salmonella is a leading cause of foodborne illness in humans worldwide [1]. Salmonella enterica subsp. enterica serovar Heidelberg is one of the most important serovars that cause infections in humans, resulting in mild diarrhea to severe systemic illness [2,3]. That is because Salmonella Heildeberg seems to be more invAsive, causing more severe disease than others serovars [4].
The virulence of Salmonella is directly related to its ability to invAde the host, to replicate within host cells, and to resist destruction by phagocytic components or plasma complement components [5]. The Salmonella invAsiveness is encoded in genes located in Salmonella Pathogenicity Island 1 (SPI-1) and the invA gene is found in this island and it is considered a marker for virulent strains of Salmonella spp [6]. The invA gene encodes several proteins involved in the Secretion Type Three System (TTSS), which is considered a molecular device that allows the bacteria to export to the interior of the host cells some proteins called “effectors” that help the bacterium to penetrate the intestinal cell, causing changes in the cytoskeleton and hence the cell framework, events that disrupt host physiology causing the appearance of symptoms that will define a more severe disease [6,7].
There has been increasing concern over the past few years regarding the worldwide emergence of multidrug-resistant phenotypes among Salmonella serovars, including S. Heidelberg. The blaCTX-M gene is usually associated to cephalosporin resistance, an antibiotic routinealy used to treat severe cases of salmonellosis in children [8]. Since the gene invA is related to pathogenesis of Salmonellosis and gene blaCTX-M to antimicrobial resistance, we hypothetized that the expression of both genes were different depending on the location of the isolation (field or slaughterhouse). Thus, the main objective of this study was to investigate, through real-time PCR (qPCR), differences on the expression of invA and blaCTXM-2 genes of S. Heidelberg isolated in the field (environmental swabs) and slaughterhouses (meat products).
Results
Real-time PCR (qPCR)
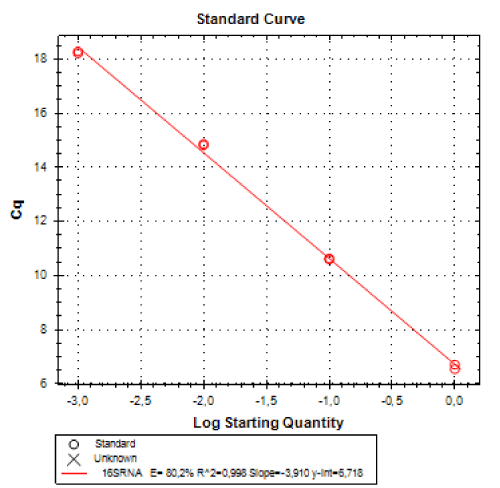
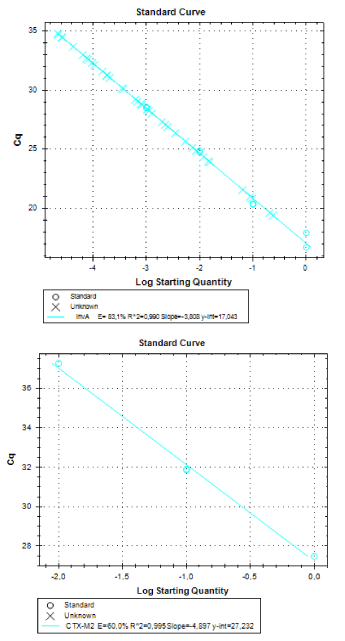
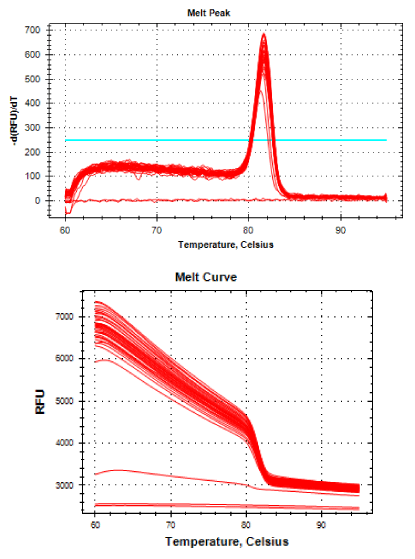
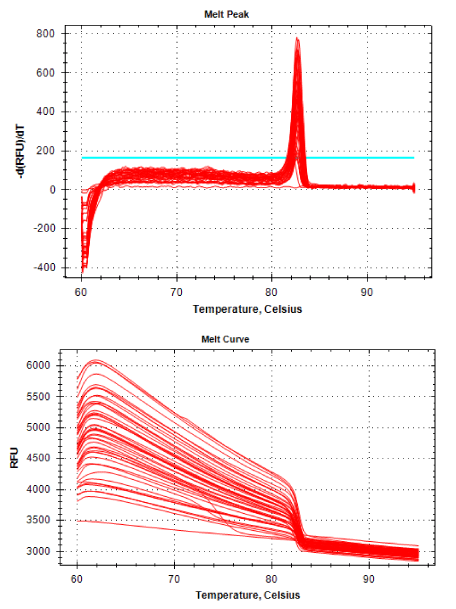
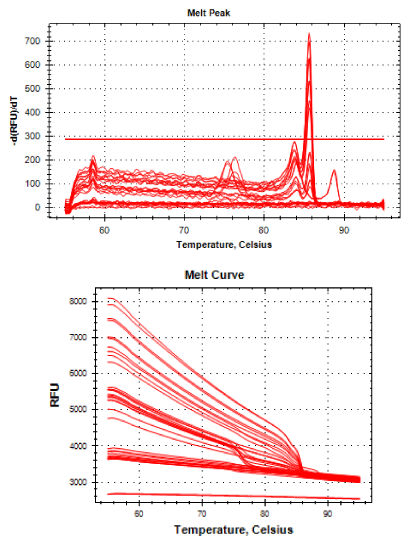
Real-time PCR was performed for 16SRNAr, invA and blaCTX-M2genes. The gene 16SRNAr was used as a control gene, and a straight line was obtained for calibration with a correlation coefficient (R2) of 0.995 and reaction Efficiency (E) of 80.2% (Figure 1). As for the genes invA and blaCTX-M2, the R2 values were 0.990 and 0.995, respectively and E values were 83.1% and 60%, respectively (Figure 2). The analyses of the melting curve and melting temperature for the 16SRNAr gene, invA and blaCTX-M2can be seen in Figure 3-5, respectively. A single peak for 16SRNAr and invA genes was identified, leading to the conclusion that there was no formation of nonspecific products, unlike blaCTX-M2gene that showed several peaks known as “shoulders”.
Table 2 shows the average values of Cq (quantification cycle) and gene expression for invA and blaCTX-M2genes for each S. Heidelberg isolate. All isolates of S. Heidelberg studied expressed invA gene. However, the amount of expression of the type III secretion system varied among isolates. Different results were observed for blaCTX-M2gene, where some isolates did not show the enzyme responsible for cefalosporin resistance. The sample number 55 (gene expression= 1:00), isolated from drag swab, was used as standard sample for the calculation of the expression of the other isolates. Among the isolates from slaugtherhouses (meat products), the sample identified as ID 53 showed invA gene expression 3.42 times greater than the control sample. Some samples from the field (drag swabs, ID= 58) showed 66.88 times higher expression than the control and it was considered the samples with greater potential of virulence.
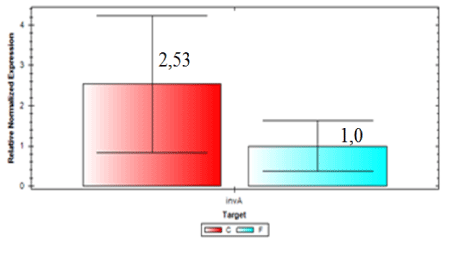
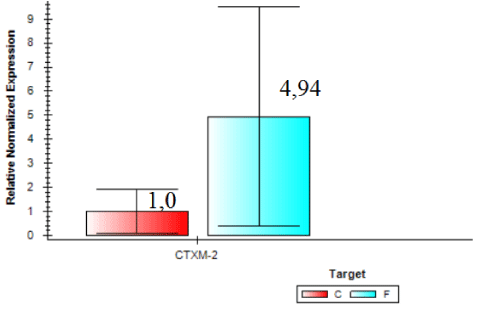
Expression of blaCTX-M2gene was higher in samples from the slaughterhouse compared to field samples (ID 54 expression was 74.84 times higher than the control sample). Field isolate (ID 56) showed gene expression 24.26 times higher than the control and were considered the sample with larger amounts of beta-lactamase enzyme expressed by blaCTX-M2. In general, invA expression was 2.53 times higher in samples collected from the field compared to samples from the slaughterhouse (Figure 6). On the other hand, Figure 7 shows that the expression of blaCTX-M2gene was 4.94 higher in samples collected at the slaughterhouse compared to field samples.
Discussion
It is possible to conclude, based on our results, that all isolates of S. Heidelberg analyzed were able to express the gene invA and that there were differences in the expression, with greater level of expression in those samples isolated from drag swabs.
Since all isolates used in these study were previously tested and considered resistants to ceftiofur, our findings regarding the expression of invA allow us to infer that the consumption of a product of poultry origin with this virulent strain of Salmonella could lead to human infection of difficult treatment [9], despite the fact that β-lactamase blaCTX-M2has not been identified by qPCR technique, since other enzymes might be responsible for the resistance .
One reason for this observed variation may be due to the great diversity of resistance genes to antibiotics encoding β-lactamases in members of the Enterobacteriaceae family [10]. According to Singh, Batish and Grover [11], the other reason for the variation in the expression of blaCTX-M2gene observed may be due to the fact that some strains lost the plasmid containing the resistance gene during storage.
The isolated ID 103 of S. Heildelberg from drag swab was shown to be even more virulent (48.10) (Table 1) and have been isolated from drag swab (field) can also become a problem for public health, as well as virulent showed expression of the gene for the enzyme blaCTX-M2(1.21) (Figure 2). The two isolates, ID 103 and ID 53 would be excellent candidates for further in vivo tests of pathogenicity. The selection of resistant bacteria may result in a more virulent, since the presence of resistance of pathogenic bacteria can lead to a delay in the administration of antimicrobial therapy may be insufficient to eliminate them [12].
Often the fact that the isolates have shown one phenotypical resistance but did not express the blaCTX-M2enzyme can be related to the presence of other β-lactamases also responsible for resistance to cephalosporins, such as blaCMY-2 and blaTEM. The resistance of β-lactamase mediated extended spectrum blaCMY-2 gene is a major mechanism of resistance to cephalosporins among strains of Salmonella enterica [12,13]. Resistance to β-lactam antibiotics, such as ceftriaxone, is correlated to an increase in the expression level of this enzyme [14]. In addition to express the invA gene, the ID 62 and 69 isolates of meat products. They showed no expression level of blaCTX-M2gene (Table 1).
Among the isolates from meat products, isolated ID 54 and 70 showed the highest levels of gene expression of blaCTX-M2enzyme, this tells us that contamination by these bacteria might have impaired treatment, as well as the presence of the gene resistance, also showed that high doses of antibiotics are necessary to eliminate the bacteria, however in vivo analysis should be conducted in the future to confirm the relationship of the expression of blaCTX-M2enzyme and resistance to ceftiofur, with possible failures in the treatment of salmonellosis. This scenario illustrates the big problem of antimicrobial resistance to public health, since all isolates were from meat products, showing the ease of transmission of these bacteria to humans by the consumption of contaminated poultry-derived products. Once infected with ESBL producing bacteria resistant to antibiotics that are commonly used for the treatment of humans, becomes more difficult and expensive therapy is often necessary hospitalizations for longer periods, it is possible that the drug is not enough to eliminate over there.
It is worth noting that the biggest problem of these superbugs are cases of complications in susceptible patients, such as those immunosuppressed. The selection of antibiotic-resistant pathogens, such as ceftiofur that have the same active principle of ceftriaxone, emphasizes once again the importance of the control of these pathogens, as these can be considered a potential source of resistant bacteria that can be transmitted to the bacteria in living beings. And if this should happen it increases the possibility of failures in the treatment of salmonellosis in children. In Figure 5 it is possible to observe a peak melting with abnormal peaks, which is called “shoulder” and is probably a consequence of the pair unspecific primers used, since the optimal size of the PCR product generated by qPCR is 80-150 bp and 486 bp not, size found. The sample 55 isolated from the ID field (drag swab) as well as having the potential for virulence (1.0), also demonstrated the expression of blaCTX-M2enzyme (1.0) (Table 1), this was the standard sample used for calibration to calculate gene expression.
Increased resistance to broad-spectrum cephalosporins (ceftiofur and ceftriaxone) from Salmonella spp isolates is of significant interest to public health. This is due to the fact that ceftriaxone is an important drug of choice for the treatment of children with severe salmonellosis. Therefore, it is possible a guess that the consumption of a product of poultry origin contaminated with a bacterium resistant to ceftiofur or even contact with an animal that is infected with this bacterium could cross-resist to ceftriaxone. As a result, the use of this agent antimicrobials in food animals is under increased scrutiny for being a potential agent responsible for the emergence and spread of resistance to ceftriaxone in Salmonella spp and other enteric pathogens [9,15]. It is important to highlight the importance of sanitary and hygienic measures in the public health system, as well as in animal husbandry production system. Thus, it is evident the need for a health program and biosecurity measures to prevent colonization and infection of animals in order to limit the spread of this epidemic bacterium [16,17].
Conclusion
All of S. Heidelberg isolates analyzed are virulent, but we observed a variation between isolates from slaughterhouses and field. By comparing the two organic groups, field isolates showed a higher expression of invA. On the other hand, by analyzing the expression of blaCTX-M2gene it was verified that isolates from slaughterhouses showed higher gene expression, and the resistance to ceftiofur could be explained by the presence of other β-lactamases and mechanisms. Further research should be conducted to better understand the mechanisms and the expression levels of these β-lactamases in the search for a solution that mitigates the prevalence of drug resistant.
Experimental procedures
RNA extraction
Molecular biology techniques were performed in the Laboratory of Molecular Biology, Immunology and Microbiology (LABMIM) of the State University of Santa Catarina (UDESC) in the West Center of High Education (CEO) in Chapecó city, Santa Catarina State, Southern Brazil. The samples submitted to RNA extraction were 18 isolates of S. Heidelberg obtained from a previous studies, well known ceftiofur resistants through the technique MIC (Minimum Inhibitory Concentration).
It should be noted that all samples were isolated in Paraná State in 2013, except the ID 104 which was isolated in 2012. First, the samples were removed from the freezer and grown in Brain Heart Infusion (BHI) for 24 hours at 37°C, followed by Brilliant Green agar for more 24 hours at 37°C. For RNA extraction, 3 to 5 colonies were isolated and inoculated again in BHI broth for 24 hours at 37°C up to a concentration of approximately 1.0×109 CFU/mL. RNA was extracted using the PureLink® RNA Mini Kit (Ambion, Life Technologies, Carlsbad, USA) and stored in liquid nitrogen (-80°C). The quality of the RNA was estimated by the OD 260/280 ratio, where ratio of 1.8 was considered optimal, indicating RNA free of proteins and other chromophores.
Reverse transcriptase
In order to sinthetize cDNA from the RNA, a Reverse Transcriptase kit (Applied Biosystems, Foster, USA) with high capacity was used. Firstly, the RNA was treated with deoxyribonuclease I enzyme - Amplification Grade (Invitrogen Life Technologies, Carlsbad).
The preparation of the RT mastermix was made up using 300 to 700 ng of RNA (10 µL), 2 µL buffer, 0.8 µL of dNTP mix (100 mM), 2 primers, 1 µL enzyme Reverse Transcriptase (RT), and nuclease-free water up to a total volume of 20 µL for each reaction. The cDNA was treated with the enzyme inhibitor RNaseOUT Recombinant Ribonuclcease™ (Invitrogen Life Technologies, Carlsbad, USA). The cDNA was stored at -15°C until use in the qPCR reaction. The preparation of the cDNA was performed in a termiciclador T100 (Bio-Rad). A Minus Reverse Transcriptase control (MRT) was used as negative control in order to assess any amount of DNA contamination in the RNA preparation.
Real-time PCR
The analysis of gene expression linked to the resistance of cephalosporins (blaCTX-M2) and the virulence gene invA was performed using real-time PCR (qPCR). As reference gene the 16SRNAr gene was used. The concentration of the primer fragment size (bp), author and reference used to assess gene expression are described in Table 2.
The qPCR reactions were performed using the kit SYBR Green PCR Master Mix (Applied Biosystems, Carlsbad, USA) according to the manufacturer’s instructions. Amplification conditions for invA and 16SRNAr genes were obtained as described by Singh and Mustafa (2013)[10] and these were: 95°C for 10 minutes, 40 cycles of denaturation 95°C for 15 seconds, annealing and extension 60°C for 45 seconds.
Amplification conditions for blaCTX-M2gene was obtained as described by Chen, et al., [18-23], which were: 95°C for 10 minutes, 40 cycles of denaturation 95°C for 30 seconds, annealing at 55 °C for 1 minute and extension at 72 °C for 1 minute and the final step was 72°C for 7 minutes. For all three genes, after 40 cycles of amplification all samples were subjected to analysis of the dissociation curve (melting curve) to confirm the absence of non-specific products and primer dimers. The samples were subjected to a gradual temperature increase of 0.1°C for 5 seconds, from 60°C up to 95°C. To determine the efficiency of the reaction and the dissociation curve reactions were optimized for the four genes evaluated. To this end, there was a pool with all samples followed by serial dilutions (pure cDNA, 1:10, 1:100 and 1:1000). Each sample was done in duplicate in specific qPCR optical plates with 96 wells, sealed with optical adhesive film, and amplified in Real Time CFX96 thermocycler (Bio-Rad). Amplification results were analyzed using the Bio-Rad CFX Manager software . A No Template Control (NTC) to omit any RNA template was used as a negative control for extraneous nucleic acid contamination.
- Majowicz SE (2010) The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 5: 882-889. Link: https://bit.ly/2Vu5d5R
- Vugia DJ, Samuel M, Farley MM, Marcus R, Shiferaw B, et al. (2004) Invasive Salmonella infections in the United States, FoodNet, 1996–1999: incidence, serotype distribution, and outcome. Clin Infect Dis 38: S149-S156. Link: https://bit.ly/2Vt0i5j
- Zhao S, White DG, Friedman SL, Glenn A, Blickenstaff K, et al. (2008) Antimicrobial resistance in Salmonella enterica serovar Heidelberg isolates from retail meats, including poultry, from 2002 to 2006. Appl Environ Microbiol 74: 6656-6662. Link: https://bit.ly/2RDMV0R
- Colla LF, Rodrigues LB, Borsoi A, Dickel EL, Nascimento VP, et al. (2012) Isolamento de Salmonella Heidelberg em diferentes pontos da tecnologia de abate de frangos de corte. Arq Inst Biol São Paulo 79: 603-606. Link: https://bit.ly/3bbvIns
- Quinn PJ, Markey BK, Carter ME, Leonard FC (2005) Microbiologia Veterinária e Doenças Infecciosas. 1. ed. Porto Alegre: Artmed 512. Link: https://bit.ly/2K7LPXe
- Vieira MAM (2009) Ilhas de patogenicidade. O mundo da saúde. São Paulo 33: 406-414.
- Grassl GA, Finlay BB (2012) Pathogenesis of enteric Salmonella infections. Curr Opin Gastroenterol 2: 22-26. Link: https://bit.ly/3abQj9U
- Catón R, Coque TM (2006) The CTX-M β-lactamase pandemic. Curr Opin Microbiol 9: 1-10. Link: https://bit.ly/2VaEhJr
- Dutil L, Irwin R, Finley R, Ng LK, Avery B, et al. (2010) Ceftiofur Resistance in Salmonella enteric Serovar Heidelberg from Chicken Meat and Humans, Canada. Emerg Infect Dis 16: 48-54. Link: https://bit.ly/3adTWvU
- Singh P, Mustapha A (2013) Multiplex TaqMan_ detection of pathogenic and multi-drug resistant Salmonella. Int J Food Microbiol 166: 213-218. Link: https://bit.ly/34ATKG1
- Singh J, Batish VK, Grover S (2012) Simultaneous detection of Listeria monocytogenes and Salmonella spp. in dairy products using real time PCR-melt curve analysis. J Food Sci Technol 49: 234-239. Link: https://bit.ly/2XA04Mo
- Biedenbach DJ, Toleman M, Walsh TR, Jones RN (2006) Analysis of Salmonella spp with resistance to extended-spectrum cephalosporins and fluoroquinolones isolated in North America and Latin America: report from the SENTRY Antimicrobial Surveillance Program (1997-2004). Diagn Microbiol Infect Dis 54: 13-21. Link: https://bit.ly/2xuULDj
- Dunne EF, Fey PD, Kludt P, Reporter R, Mostashari F, et al. (2000) Emergence of domestically acquired ceftriaxone resistant Salmonella infections associated with AmpC β-lactamase. JAMA 284: 3151-3156. Link: https://bit.ly/2yZUeJT
- Whichard JM, Gay K, Stevenson JE, Joyce KJ, Cooper KL, et al. (2007) Human Salmonella and concurrent decreased susceptibility to quinolones and extended-spectrum cephalosporins. Emerg Infect Dis 13: 1681-1688.
- Foley SL, Lynne AM (2007) Food animal-associated Salmonella challenges: pathogenicity and antimicrobial resistance. J Anim Sci 86: 173-187. Link: https://bit.ly/3cdHIoq
- Bond R, Loeffler A (2012) What's happened to Staphylococcus intermedius Taxonomic revision and emergence of multi-drug resistance. J Small Anim Pract 53: 147-154. Link: https://bit.ly/34JOtvS
- Frye JG, Jackson CR (2013) Genetic mechanisms of antimicrobial resistance identified in Salmonella enterica, Escherichia coli, and Enteroccocus spp. Isolated from U.S. food animals. Front Microbiol 4: 1-22. Link: https://bit.ly/2xuUkJ9
- Chen S, Zhao S, White DG, Schroeder CM, Lu R, et al. (2004) Characterization of Multiple-Antimicrobial-Resistant SalmonellaSerovars Isolated from Retail Meats. Appl Environ Microbiol 70: 1-7. Link: https://bit.ly/2XDmMmW
- Chiu CH, Su LH, Chu CH, Wang MH, Yeh CM, et al. (2006) Detection of multidrug-resistant Salmonella enteric serovar Typhimurium phage types DT102, DT104, and U302 by multiplex PCR. J Clin Microbiol 44: 2354-2358. Link: https://bit.ly/3cohH65
- Suarez C, Gudiol F (2009) Beta-lactam antibiotics. Enferm Infecc Microbiol Clin 27: 116-129. Link: https://bit.ly/3bczexF
- Sturenburg E, Sobottka L, Laufs R, Mack D (2005) Evaluation of a new screen agar plate for detection and presumptive identification of Enterobacteriaceae producing extended-spectrum betalactamases. Diagn Microbiol Infect Dis 51: 51-55. Link: https://bit.ly/3eq3GX4
- Tenover FC (2006) Mechanisms of antimicrobial resistance in bacteria. Am J Infect Control 119: 3-10. Link: https://bit.ly/2Vt2vxD
- Utiyama CE, Oetting LL, Giani PA, Ruiz SdU, Miyada VS (2006) Efeitos de antimicrobianos, prebioticos, probioticos e extratos vegetais sobre a microbiota intestinal, a requencia de diarreia e o desempenho de leitões recem desmamados. Revista Brasileira de Zootecnia 35: 2359-2367. Link: https://bit.ly/3ci0uLu
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
