Annals of Molecular and Genetic Medicine
Exome sequencing reveals a homozygous frameshift variant in CAPN3 in a Tunisian patient with a neuromuscular disorder
Hajer Foddha1, Go Hun Seo2, Hane Lee2, Firas Zemzem3, Ines Naouar1, Amel Boughammoura4, Moez Gribaa5 and Amel Haj Khelil1,3*
2Division of Medical Genetics, 3billion Inc., Seoul, South Korea
3Superior Institute of Biotechnology, University of Monastir, Tunisia
4Free practice Doctor of Neurology at Monastir, Tunisia
5Laboratory of Cytogenetics, Molecular Genetics and Human Reproduction Biology CHU Farhat Hached, Sousse, Tunisia
Cite this as
Foddha H, Seo GH, Lee H, Zemzem F, Khelil AH, et al. (2022) Exome sequencing reveals a homozygous frameshift variant in CAPN3 in a Tunisian patient with a neuromuscular disorder. Ann Mol Genet Med 6(1): 001-004. DOI: 10.17352/amgm.000010Copyright License
© 2022 Foddha H, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Muscular dystrophy (MD) is a heterogeneous group of diseases that cause progressive weakness and loss of muscle mass. Specific signs and symptoms begin at different ages and in different muscle groups, depending on the type of muscular dystrophy. We report here a case of a Tunisian patient suffering from a neuromuscular disorder, highly suspicious of a Limb-Girdle Muscular Dystrophy (LGMD). Exome Sequencing revealed a one-base deletion variant in exon 4 of the CAPN3 gene. CAPN3 is associated with a recessive form of LGMD, also known as Muscular Dystrophy, Limb-Girdle, Type 2a (LGMD2A). The variant was shown to segregate with the disease in the family. The identification of the molecular defect in this family provided a rapid genotyping for the sister and an accurate diagnosis for the patient, allowing appropriate clinical management at an early stage of the disease.
Abbreviations
CAPN3: Calpain-3 protein; dbSNP: SNP database; DMD: Duchenne Muscular Dystrophy; LGMD2A: Limb-Girdle Muscular Dystrophy type 2A; LGMDR1: Limb-Girdle Muscular Dystrophy Recessive 1; MD: Muscular dystrophy; ES: Exome Sequencing
Introduction
Muscular Dystrophy (MD) is a group of diseases that cause progressive weakness and loss of muscle mass. In muscular dystrophies, pathogenic variants interfere with the production of proteins needed to form healthy muscle. The main sign in all MDs is progressive muscle weakness. Specific signs and symptoms begin at different ages and in different muscle groups, depending on the type of MD. In effect, MDs have a wide range of clinical forms (Duchenne, Becker, Limb-girdle, Myotonic, Congenital and Facioscapulohumeral). The most common type is Duchenne Muscular dystrophy (DMD). The Limb-girdle form (LGMD) is the fourth most common form with an estimated prevalence of about 2 in every 100,000 individuals [1,2,3]. LGMDs are classified by the respective inheritance patterns: LGMD1 for autosomal dominant and LGMD2 for autosomal recessive. Dozens of different genes, when mutated, have been shown to cause different LGMD1 and LGMD2 types. Depending on the genes involved, each type of LGMD is further subdivided into several subtypes (LGMD1A, LGMD1B,…, LGMD1H) for dominant forms and (LGMD2A, LGMD2B,…., LGMD2Y) for recessive forms. LGMD2A is the most common type worldwide, accounting for 15-40% of all LGMD cases [4]. As there are still many patients with the symptom of LGMD for whom the molecular diagnosis is not established even after receiving a genetic test, it is suspected that there are novel genes yet to be discovered for LGMDs.
Here, we report a 24-year-old patient with multiple signs of LGMD that started to manifest at the age of 10. A thorough clinical evaluation has been performed but it could not specify which type of LGMD the patient has, requiring a molecular diagnosis.
Material and methods
Patient
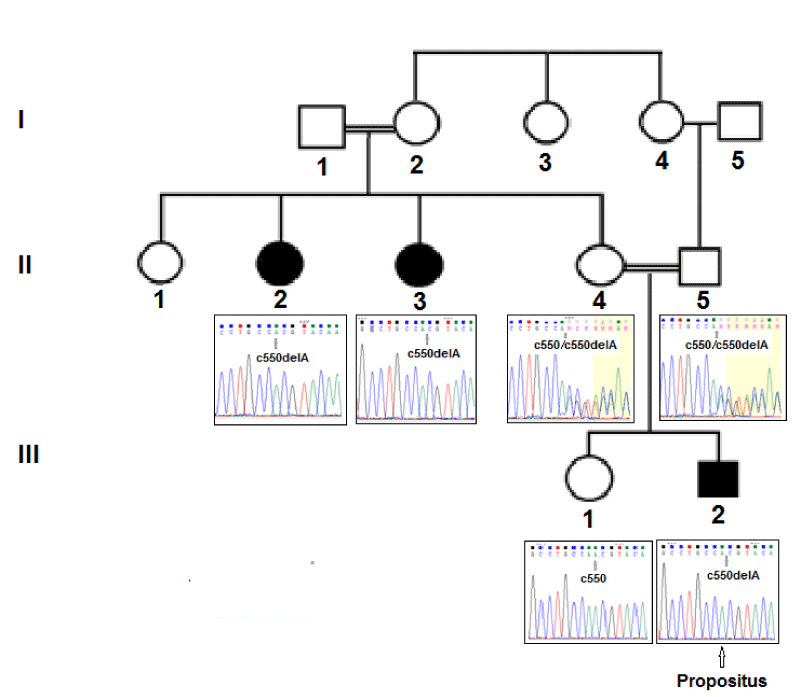
The proband was a 24-year-old patient born from a first-degree consanguineous marriage, suffering from LGMD. His two parents did not show any symptoms of the disease. However, two maternal aunts shared the same symptoms as the proband (Figure 1). The pedigree supports an autosomal recessive mode of inheritance that corresponds to the LGMD2 type. Since his schooling, the patient has had difficulty running, climbing stairs and getting up from the ground. These gradually worsened, reducing his physical activities. His motor deficit was proximal, affecting both girdles, with moderate atrophy of the hip and para-vertebral muscles. The muscle biopsy confirmed progressive muscular dystrophy. He also presented a worsening of dandian walking disorders, sometimes needing a wheelchair for long distances.
Methods
Genomic DNA was extracted from white blood cells using a standard phenol/chloroform method and sent to 3billion, Inc for exome sequencing (ES). ES was performed following 3billion’s standard protocol: exome capture was performed by xGen Exome Research Panel v2 (Integrated DNA Technologies, Coralville, Iowa, USA) and sequencing was run on Illumina NovaSeq 6000 (Illumina, Inc, San Diego, CA, USA) generating 150 bp paired-end reads. Sequence data analysis was performed according to previously published protocol [5]. Mean depth of ~225x was generated and ~99 % of the targeted regions were covered at >=20X. Variant analysis was performed using 3 billion’s EVIDENCE [5] to annotate, filter and classify the variants based on the American College of Medical Genetics and Genomics (ACMG) guideline [6] and prioritize by the symptom similarity score. Finally, the variant was interpreted in the context of the patient’s phenotype by 3 billion’s Medical Geneticists. Sanger sequencing was performed on the proband and the family members using the ABI PRISM 3100 Genetic Analyzer and the BigDye Terminator Cycle Sequencing Kit from ThermoFisher scientific.
Results
Genetic analysis
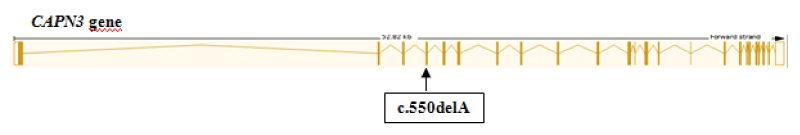
Exome sequencing identified a one-base deletion variant 15-42680000-CA-C (GRCh37) in exon 4 of the CAPN3 gene on chromosome 15. This variant NM_000070.2:c.550delA (Figure 1) is predicted to result in a frameshift (NP_000061.1:p.Thr184ArgfsTer36), leading to early termination of the protein. CAPN3 gene encodes calpain-3, a protein that constitutes a muscle-specific calcium-dependent intracellular protease. CAPN3 is associated with both autosomal dominant LGMD4 (also known as LGMD1I) and autosomal recessive LGMD1 (also known as LGMD2A). The variant is a well-established pathogenic variant that has been reported in multiple other patients with a similar phenotype (ClinVar ID: VCV000017621). There were no additional variants that could explain the proband’s phenotype.
Segregation analysis
Sanger sequencing confirmed the homozygous c.550delA variant in the proband (III2). His two similarly-affected aunts (II2; II3) were also homozygous and the two unaffected parents were each heterozygous (II4; II5) for the same variant. The unaffected sister (III1)(Figure 2) was homozygous for the reference allele.
Discussion
The autosomal recessive Limb-girdle muscular dystrophy type 2A (LGMD2A), also known as calpainopathy or LGMDR1 [7] (limb-girdle muscular dystrophy recessive 1) is caused by homozygous or compound heterozygous variants in the calpain 3 (CAPN3) gene encoding for the proteolytic enzyme calpain 3 (CAPN3) [8]. LGMD2A is characterized by selective atrophy and progressive weakness of the shoulder and proximal limb and girdle muscles that usually appears in children and young adults. It results in loss of ambulation within 10 to 20 years after the disease onset in most patients [9,10].
Since the discovery of CAPN3 as the gene responsible for LGMD2A, various studies performed in animal models and human samples have shown that CAPN3 deficiency is associated with different features in the skeletal muscle such as oxidative damage, Ca2+ dysregulation, sarcomere disorganization, mitochondrial abnormalities, abnormal muscle adaptation and impaired muscle regeneration, which together would lead to inflammation, necrosis, fibrosis, atrophy and progressive muscle degeneration, characteristic of LGMD2A [9].
Even though the c.550delA variant is a well-established pathogenic variant, it is found in the population of individuals who are carriers. In the gnomAD v2.1.1 (https://gnomad.broadinstitute.org/) dataset, it is observed in 65 individuals as heterozygous with an allele frequency of 0.000228. Most of these individuals are of European descent (allele frequency of 0.000648) and only 1 out of 24,964 African population is heterozygous for the variant (allele frequency of 0.0000401). Being consistent with this observation, this variant is most frequent in the patients from Eastern and Central Europe [11-17]. For example, in Latvia, it had been described with an allele frequency of 0.13 when compared to the controls group (0.001) [11] while it has never been described in Tunisian and African patients, making this case report significant. This variant in our patient’s family could come from a common ancestor who probably had migrated from Eastern Europe (especially from Turkey where the variant has already been described)) to Tunisia during the Ottoman Empire. Indeed, the Ottomans settled in Tunisia during the 16th century and mixed with the endogenous population. Further haplotypic analysis may clarify this hypothesis of the c.550delA origin in North Africa.
This variant is expected to cause a loss of normal calpain-3 protein function via nonsense-mediated mRNA decay although there is no functional evidence reported.
The only variants previously identified on the CAPN3 gene in LGMD2A Tunisian patients are c.1536+1G>T in the donor splice site of the 12th intron [18] and a missense mutation (c.T1681C/p.Y561H) in the 13th exon [19].
Since the pathophysiological mechanisms involved in LGMDR1 remain mostly unknown, there is no effective treatment or cure for this disease. However, medications and therapy can help manage symptoms and slow the course of the disease and therefore it is important to identify the molecular diagnosis as early as possible. Performing further analyses at the biochemical level such as western blot using antibodies directed against the calpain 3 protein and at the molecular level using the real-time PCR technique to quantify the expression of the CAPN3 gene in patients are in the work. A functional study by site-directed mutagenesis of the c.550delA variant can also be envisaged.
Conclusion
Exome sequencing allowed to end of the “diagnostic odyssey” in this family, providing opportunities for early clinical management and genetic counseling for the family members.
The authors would like to thank 3billion’s “Rare Disease Genetic Testing” program for the collaboration and the patient’s family for their participation in the study.
- Understanding Neuromuscular Disease Care. IQVIA Institute. 2018.
- Narayanaswami P, Weiss M, Selcen D, David W, Raynor E, Carter G, Wicklund M, Barohn RJ, Ensrud E, Griggs RC, Gronseth G, Amato AA; Guideline Development Subcommittee of the American Academy of Neurology; Practice Issues Review Panel of the American Association of Neuromuscular & Electrodiagnostic Medicine. Evidence-based guideline summary: diagnosis and treatment of limb-girdle and distal dystrophies: report of the guideline development subcommittee of the American Academy of Neurology and the practice issues review panel of the American Association of Neuromuscular & Electrodiagnostic Medicine. Neurology. 2014 Oct 14;83(16):1453-63. doi: 10.1212/WNL.0000000000000892. PMID: 25313375; PMCID: PMC4206155.
- Wicklund MP. Limb-Girdle Muscular Dystrophies. in Encyclopedia of the Neurological Sciences. 2014; 890-96. Elsevier.
- Limb-Girdle Muscular Dystrophy (LGMD). Muscular Dystrophy Association. (//www.mda.org/disease/limb-girdle-muscular-dystrophy)
- Seo GH, Kim T, Choi IH, Park JY, Lee J, Kim S, Won DG, Oh A, Lee Y, Choi J, Lee H, Kang HG, Cho HY, Cho MH, Kim YJ, Yoon YH, Eun BL, Desnick RJ, Keum C, Lee BH. Diagnostic yield and clinical utility of whole exome sequencing using an automated variant prioritization system, EVIDENCE. Clin Genet. 2020 Dec;98(6):562-570. doi: 10.1111/cge.13848. Epub 2020 Sep 17. PMID: 32901917; PMCID: PMC7756481.
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015 May;17(5):405-24. doi: 10.1038/gim.2015.30. Epub 2015 Mar 5. PMID: 25741868; PMCID: PMC4544753.
- Straub V, Murphy A, Udd B; LGMD workshop study group. 229th ENMC international workshop: Limb girdle muscular dystrophies - Nomenclature and reformed classification Naarden, the Netherlands, 17-19 March 2017. Neuromuscul Disord. 2018 Aug;28(8):702-710. doi: 10.1016/j.nmd.2018.05.007. Epub 2018 May 24. PMID: 30055862.
- Richard I, Broux O, Allamand V, Fougerousse F, Chiannilkulchai N, Bourg N, Brenguier L, Devaud C, Pasturaud P, Roudaut C, et al. Mutations in the proteolytic enzyme calpain 3 cause limb-girdle muscular dystrophy type 2A. Cell. 1995 Apr 7;81(1):27-40. doi: 10.1016/0092-8674(95)90368-2. PMID: 7720071.
- Wang CH, Liang WC, Minami N, Nishino I, Jong YJ. Limb-girdle muscular dystrophy type 2a with mutation in CAPN3: the first report in Taiwan. Pediatr Neonatol. 2015 Feb;56(1):62-5. doi: 10.1016/j.pedneo.2013.01.018. Epub 2013 Mar 7. PMID: 23597518.
- Lasa-Elgarresta J, Mosqueira-Martín L, Naldaiz-Gastesi N, Sáenz A, López de Munain A, Vallejo-Illarramendi A. Calcium Mechanisms in Limb-Girdle Muscular Dystrophy with CAPN3 Mutations. Int J Mol Sci. 2019 Sep 13;20(18):4548. doi: 10.3390/ijms20184548. PMID: 31540302; PMCID: PMC6770289.
- Inashkina I, Jankevics E, Stavusis J, Vasiljeva I, Viksne K, Micule I, Strautmanis J, Naudina MS, Cimbalistiene L, Kucinskas V, Krumina A, Utkus A, Burnyte B, Matuleviciene A, Lace B. Robust genotyping tool for autosomal recessive type of limb-girdle muscular dystrophies. BMC Musculoskelet Disord. 2016 May 4;17:200. doi: 10.1186/s12891-016-1058-z. PMID: 27142102; PMCID: PMC4855345.
- Sáenz A, Leturcq F, Cobo AM, Poza JJ, Ferrer X, Otaegui D, Camaño P, Urtasun M, Vílchez J, Gutiérrez-Rivas E, Emparanza J, Merlini L, Paisán C, Goicoechea M, Blázquez L, Eymard B, Lochmuller H, Walter M, Bonnemann C, Figarella-Branger D, Kaplan JC, Urtizberea JA, Martí-Massó JF, López de Munain A. LGMD2A: genotype-phenotype correlations based on a large mutational survey on the calpain 3 gene. Brain. 2005 Apr;128(Pt 4):732-42. doi: 10.1093/brain/awh408. Epub 2005 Feb 2. PMID: 15689361.
- Hanisch F, Müller CR, Grimm D, Xue L, Traufeller K, Merkenschlager A, Zierz S, Deschauer M. Frequency of calpain-3 c.550delA mutation in limb girdle muscular dystrophy type 2 and isolated hyperCKemia in German patients. Clin Neuropathol. 2007 Jul-Aug;26(4):157-63. doi: 10.5414/npp26157. PMID: 17702496.
- Pogoda TV, Krakhmaleva IN, Lipatova NA, Shakhovskaya NI, Shishkin SS, Limborska SA. High incidence of 550delA mutation of CAPN3 in LGMD2 patients from Russia. Hum Mutat. 2000 Mar;15(3):295. doi: 10.1002/(SICI)1098-1004(200003)15:3<295::AID-HUMU15>3.0.CO;2-8. PMID: 10679950.
- Canki-Klain N, Milic A, Kovac B, Trlaja A, Grgicevic D, Zurak N, Fardeau M, Leturcq F, Kaplan JC, Urtizberea JA, Politano L, Piluso G, Feingold J. Prevalence of the 550delA mutation in calpainopathy (LGMD 2A) in Croatia. Am J Med Genet A. 2004 Mar 1;125A(2):152-6. doi: 10.1002/ajmg.a.20408. PMID: 14981715.
- Peric S, Stevanovic J, Johnson K, Kosac A, Peric M, Brankovic M, Marjanovic A, Jankovic M, Banko B, Milenkovic S, Durdic M, Bozovic I, Glumac JN, Lavrnic D, Maksimovic R, Milic-Rasic V, Rakocevic-Stojanovic V. Phenotypic and genetic spectrum of patients with limb-girdle muscular dystrophy type 2A from Serbia. Acta Myol. 2019 Sep 1;38(3):163-171. PMID: 31788660; PMCID: PMC6859413.
- Topaloğlu H, Dinçer P, Richard I, Akçören Z, Alehan D, Ozme S, Cağlar M, Karaduman A, Urtizberea JA, Beckmann JS. Calpain-3 deficiency causes a mild muscular dystrophy in childhood. Neuropediatrics. 1997 Aug;28(4):212-6. doi: 10.1055/s-2007-973702. PMID: 9309711.
- Hadj Salem I, Kamoun F, Louhichi N, Rouis S, Mziou M, Fendri-Kriaa N, Makni-Ayadi F, Triki C, Fakhfakh F. Mutations in LAMA2 and CAPN3 genes associated with genetic and phenotypic heterogeneities within a single consanguineous family involving both congenital and progressive muscular dystrophies. Biosci Rep. 2011 Apr;31(2):125-35. doi: 10.1042/BSR20100026. PMID: 20477750.
- Rekik S, Sakka S, Ben Romdhan S, Farhat N, Baba Amer Y, Lehkim L, Authier FJ, Mhiri C. Novel Missense CAPN3 Mutation Responsible for Adult-Onset Limb Girdle Muscular Dystrophy with Calves Hypertrophy. J Mol Neurosci. 2019 Dec;69(4):563-569. doi: 10.1007/s12031-019-01383-z. Epub 2019 Aug 13. PMID: 31410652.
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
