Annals of Molecular and Genetic Medicine
The expression of DLGAP5 associate with progression and prognosis in glioma
Kangjie Du1,2, Yu Zhang1,2, Mengyao He1,2, Yalin Lu3, Xingjie Chen1,2, Hao Yu1,2 and Qiang Huang1,2*
2Tianjin Key Laboratory of Injuries, Variations and Regeneration of Nervous System, Tianjin, China
3Department of Neurosurgery, Henan Provincial People’s Hospital, Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, China
Cite this as
Du K, Zhang Y, He M, Lu Y, Chen X, et al. (2022) The expression of DLGAP5 associate with progression and prognosis in glioma. Ann Mol Genet Med 6(1): 005-016. DOI: 10.17352/amgm.000011Copyright License
© 2022 Du K, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Glioma is the most common primary malignant tumor of the central nervous system and is related to poor clinical outcomes. At present, the standard treatment of glioma in clinical practice is to maximally remove the focus on the premise of protecting the neurological function, supplemented by postoperative chemotherapy and radiotherapy. However, after standard treatment, the prognosis of glioma patients is still not satisfactory. DLGAP5 has been shown to play an important role in the occurrence and progression of various tumors. This study examined the expression of DLGAP5 in glioma samples and its significance in predicting the prognosis of glioma patients. TCGA and CGGA datasets were used to explore the difference in DLGAP5 expression between glioma and normal central nervous system tissues and to investigate the prognostic value of DLGAP5 expression in glioma patients. The cBioPortal online analysis website was used to explore the gene mutations of DLGAP5 expressing in glioma. The String database and GEPIA online analysis website were used to perform Enrichment Analysis to explore the molecular mechanism of DLGAP5.In this study, we observed that DLGAP5 expression was upregulated in glioma tissues compared with normal CNS tissues and was negatively associated with the prognosis of glioma patients. DLGAP5 may affect the occurrence and progression of glioma mainly through gene mutation, gene amplification, and gene depth deletion. At the same time, the molecular inhibition of DLGAP5 may be related to the cell cycle and the p53 pathway. In conclusion, our findings suggested that DLGAP5 may be a therapeutic target and an independent prognostic indicator for glioma.
Abbreviations
WHO: World Health Organization; DLGAP5: Discs Large Homolog Associated Protein 5; TIMER: Tumor Immune Estimation Resource; CGGA: Chinese Glioma Genome Atlas; TCGA: The Cancer Genome Atlas; GEPIA: Gene Expression Profiling Interactive Analysis; TMB: Tumor Mutation Burden; MSI: Microsatellite Instability; CAFs: Tumor-Associated Fibroblasts; ICP: Immune Checkpoints; PPI: Protein-Protein Interaction network; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; BP: Biological Process; CC: Cellular Component; MF: Molecular Function
Background
Glioma is the most common primary malignant tumor in the central nervous system [1]. According to the 2021 World Health Organization Classification of central nervous system tumors, glioma can be divided into 4 grades, of which WHO grand1 and WHO grand 2 are low-grade gliomas, WHO grand 3 and grand 4 are high-grade gliomas [2]. At present, the standard treatment of glioma in clinical practice is to maximally remove the focus on the premise of protecting the neurological function, supplemented by postoperative chemotherapy and radiotherapy. Because of the rapid growth and invasiveness of glioma, the clinical treatment of glioma is complicated and difficult [3]. The median survival time of high-grade glioma patients is 15 months, and the 5-year survival rate is less than 5% [4]. It is crucial for glioma patients to find a new diagnostic method and more effective treatments. In recent years, with the development of molecular biology, we have a better understanding of molecular changes in gliomas [5]. Through previous studies and the success of other clinical targeted therapies for other tumors, the intervention of molecules closely related to the occurrence and prognosis of glioma may have a positive impact on the diagnosis and prognosis of patients with glioma.
Discs Large Homolog Associated Protein 5 (DLGAP5), also known as DLGAP7 or Hepatoma Up-Regulated Protein (HURP), is a mitotic spindle protein encoded by the DLGAP5 gene in humans [6]. DLGAP5, as a kinetochore protein, stabilizes K fibers and promotes chromosome aggregation by regulating the localization and dynamics of kif18A at the positive end of kinetochore microtubules, thus stabilizing the microtubules near chromosomes [7-9]. DLGAP5 controls spindle dynamics and stabilizes in cells through phosphorylation by Aurora kinase. It also plays an important role in centrosome formation and chromosome separation, thus regulating spindle formation [10-12]. Previous studies have found that DLGAP5 is closely related to the occurrence and progression of tumors. High expression of DLGAP5 is common in colorectal cancers with poor overall survival. By down-regulating the expression of the DLGAP5 gene, it significantly reduced the invasion and migration potential of colorectal cancer [13]. Previous studies have found that DLGAP5 expression promotes the development of NSCLC [14]. DLGAP5 is overexpressed in hepatocellular carcinoma, and DLGAP5 silencing inhibits the cycle and proliferation of hepatocellular carcinoma cells [15]. In one study, the expression of DLGAP5 in endometrial cancer tissues was significantly higher than that in normal endometrial tissues, and the expression level of DLGAP5 was negatively correlated with the prognosis of patients with endometrial cancer [16]. Studies have shown that DLGAP5 is a potential new molecular biomarker for the diagnosis and prognosis of glioma [17]. DLGAP5 plays an important role in the occurrence, progression, and prognosis of glioma, colorectal cancer, lung cancer, hepatocellular carcinoma, and endometrial carcinoma. However, the role of DLGAP5 in the development and progression of glioma is rarely studied in the basic and clinical studies of glioma.
Although studies have proved that DLGAP5 may be a new molecular biomarker for the diagnosis and prognosis of glioma, the mechanism through which DLGAP5 affects the occurrence and progression of glioma remains unclear. In this study, we performed integrated bioinformatics analysis based on gene expression profiles in relevant databases to explore the possible mechanisms of DLGAP5. We used TCGA, CGGA database, and related online bioanalysis websites to investigate the differential expression of DLGAP5 in glioma tissues and normal central nervous system tissues and its prognostic impact on glioma patients. This study also investigated the correlation between DLGAP5 gene mutation and immune cell infiltration. In addition, we performed GSEA (Gene ensemble Enrichment analysis) to explore the mechanism of DLGAP5 action.
Materials and methods
TIMER2
TIMER (Tumor Immune Estimation Resource) is a comprehensive resource system for the analysis of immune infiltration in different cancer types. It can fully explore the immunological, clinical, and genomic aspects of tumors. The Gene-DE module in TIMER2.0 can investigate the expression differences of any corresponding gene between the tumor and adjacent normal tissues in all TCGA tumors. The distribution of gene expression levels was represented by boxplots. In this study, we used the gene-DE plate in TIMER2.0 to investigate the differential expression of DLGAP5 in different tumor tissues, especially glioma and corresponding normal tissues. Meanwhile, the TIMER2 online database was used to study the correlation between DLGAP5 and the level of immune cell infiltration in glioma [18-20].
CGGA
The Chinese Glioma Genome Atlas (CGGA) database, a web-based application focused on brain tumor data storage and analysis, explores more than 2000 datasets of brain tumor samples from a Chinese cohort. This analysis tool can scan DNA mutation profiles, mRNA/microRNA expression profiles, and methylation profiles, and perform correlation and survival analyses for specific glioma subtypes. We used different data sources in the CGGA database (data ID: mRNAseq-325, mRNAseq-693, mRNA-array-301) to investigate the expression differences of DLGAP5 in glioma of different WHO grades. At the same time, the CGGA database was used to explore the correlation between the expression level of DLGAP5 and the prognosis of glioma patients [21-23].
TCGA
The Cancer Genome Atlas (TCGA) is a publicly funded project that aims to catalog and discover major cancer-causing genomic alterations to create a comprehensive “atlas” of cancer genomic profiles. So far, TCGA researchers have analyzed large cohorts of over 30 human tumors through large-scale genome sequencing and integrated multi-dimensional analyses. Studies of individual cancer types, as well as comprehensive pan-cancer analyses, have extended current knowledge of tumorigenesis. A major goal of the project was to provide publicly available datasets to help improve diagnostic methods, and treatment standards, and finally to prevent cancer [24]. In this study, The TCGA database was used to analyze the expression level of DLGAP5 in glioma and its correlation with the prognosis of glioma patients. Meanwhile, the TCGA database was used to explore the correlation between DLGAP5 and TMB, and MSI.
cBioPortal
The cBioPortal for Cancer Genomics is a comprehensive open network platform based on the TCGA database, which integrates data mining, data integration, and visualization. The portal collected records from 147cancer studies and analyzed 31 cancers, including more than 21000 samples. The variation of DLGAP5 in glioma was studied by using the “Cancer Types Summary” and “Mutations” sections of this network platform. In the current tumor research, tumor immunotherapy has received attention from basic and clinical research [25,26].
String
The String database is a database that searches for known protein interactions and predicts protein-protein interactions. Firstly, 50 genes related to DLGAP5 expression were obtained by using the Protein by name module in the sting database. Then the protein-protein interaction network is obtained by setting the Meaning of network edges selection [evidence], Active interaction sources selection [Experiments], Minimum required interaction score selection [low confidence (0.150)], Max number of interactors selection [no more than 50 interactors] and so on [27-29]. The String database was used to explore DLGAP5 protein interactions and predict protein-protein interactions.
GEPIA
GEPIA (Gene Expression Profiling Interactive Analysis) is a web server for cancer and normal gene expression and interaction analysis. GEPIA2.0 is a version that adds functionality to it. By using the Similar Genes Detection module of GEPIA2 to obtain the first 100 genes related to DLGAP5 expression, we took the first five genes, used the Correlation Analysis module to analyze the correlation between the expression of the first five genes and DLGAP5 expression, and obtained the related scatter plot [30].
DAVID
DAVID (The Database for Annotation, Visualization, and Integrated Discovery) provides a comprehensive set of functional annotation tools for investigators to understand the biological meaning behind large lists of genes. These tools are powered by the comprehensive DAVID Knowledgebase built upon the DAVID Gene concept which pulls together multiple sources of functional annotations. For any given gene list, DAVID tools are able to identify enriched biological themes, particularly GO terms and discover enriched functional-related gene groups [31]. The differentially expressed genes (DEGs) from differential expression analysis that meet the conditions were used for enrichment analysis, including the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) enrichment analysis. The DAVID was used to conduct GO enrichment analysis for the candidate target protein obtained after network merging. We could use DAVID to visualize the enrichment of BP, MF, CC, and pathways (p < 0.05). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was conducted to explore biological pathways where relevant proteins were covered. DAVID and R language ggplot2 package were used to enrich analysis visualization and analyze the possible mechanism of actions, [32].
Results
The expression level of DLGAP5 in glioma
Genes are often involved in the occurrence and progression of tumors. In order to verify whether DLGAP5 is a glioma-promoting gene or a tumor suppressor gene, we used a variety of databases to analyze its expression level in gliomas and its difference with normal tissues.
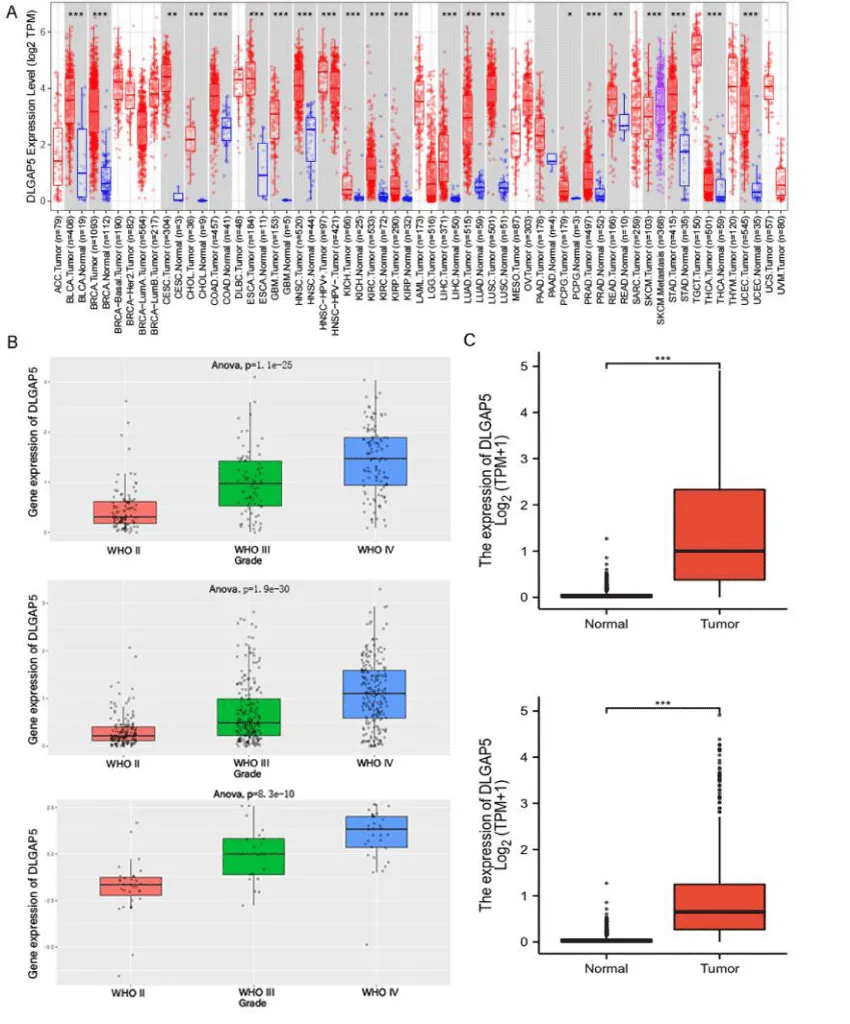
In this study, the TIMER2.0 database was used to explore and analyze the differences in DLGAP5 expression in different tumors, especially in glioma. Then CGGA and TCGA were used to verify their expression level again. Figure 1 can see that DLGAP5 is highly expressed in a variety of tumors. Among the 153 glioblastoma samples from the TIMER2.0 database, DLGAP5 was highly expressed in tumor cells, while DLGAP5 was low in 5 normal glioblastoma tissues (p < 0.001). DLGAP5 is also highly expressed in Low-grade gliomas (Figure 1A). In order to further study the expression level of DLGAP5 in glioma, we used different data sources in the CGGA database (data ID: mRNAseq-325, mRNAseq-693, mRNA-array-301) to analyze the differences of DLGAP5 expression in different WHO grades of glioma. As shown in Figure 1B, the expression level of DLGAP5 is the highest in WHO grade 4 glioma, the lowest in WHO grade 2 glioma, and in the middle of WHO grade 3 glioma. In this study, we verified the results of TIMER by extracting data from the TCGA database. The expression of DLGAP5 was up-regulated in glioblastoma and low-grade glioma, but not in normal tissues, (Figure 1C).
From the expression level of DLGAP5 in glioma, it can be concluded that DLGAP5 is highly expressed in glioblastoma and Low-grade glioma, and there is a significant statistical difference between glioblastoma and normal tissue. The results of this data showed that the expression level of DLGAP5 was significantly different in different grades of gliomas. The expression level of DLGAP5 in high-grade gliomas is higher than that in low-grade gliomas.
Prognosis of glioma patients with DLGAP5 expression
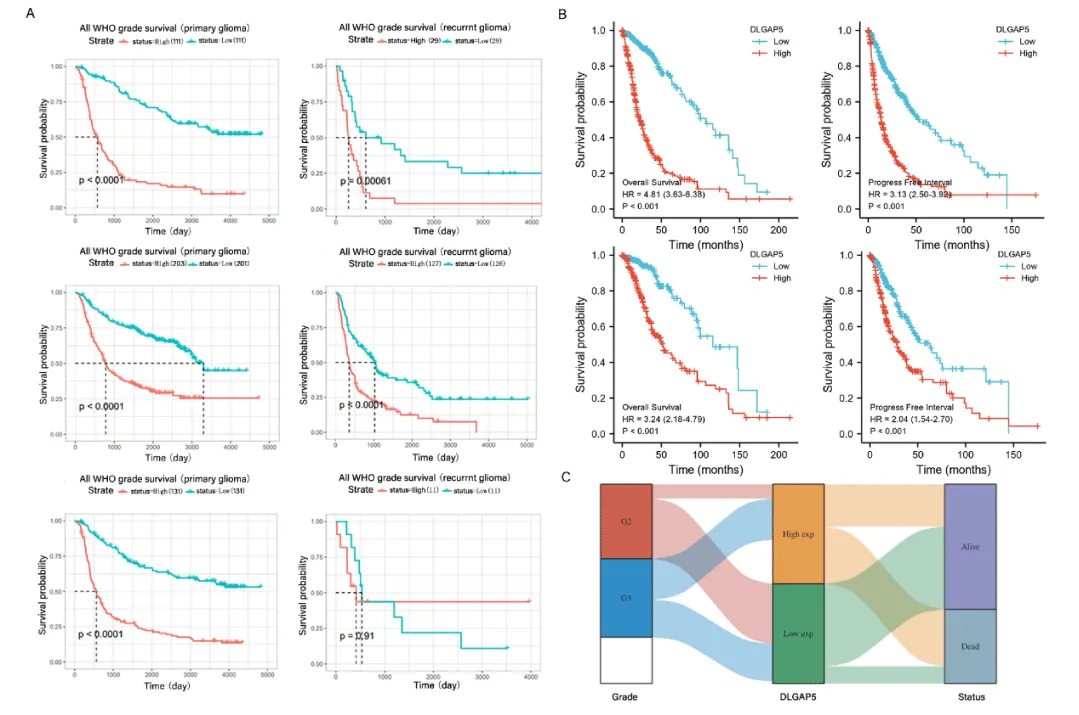
In this study, we used multiple bioinformatics databases to investigate the effect of DLGAP5 expression level on the prognosis of glioma patients. First of all, this study used three different data sources in the CGGA database (data ID: mRNAseq-325, mRNAseq-693, mRNA-array-301) for analysis. The samples were divided into the DLGAP5 high expression group and low expression group, and the effect of DLGAP5 expression on the prognosis of primary glioma and recurrent glioma was studied respectively. We used data from the CGGA database to explore the effect of DLGAP5 expression level on the prognosis of glioma patients in primary glioma and recurrent glioma. The data show that in primary glioma, the survival probability of the group with high expression of DLGAP5 was lower than that of the group with low expression (p < 0.0001). In recurrent glioma, the data from the CGGA database (data ID: mRNAseq-325 and mRNAseq-693) showed that the survival probability of the high-expression group of DLGAP5 was lower than that of the low-expression group. The data obtained from the CGGA database (data ID: mRNA-ARRAY-301) had no statistically significant results. Our team thought that the small sample size made it impossible to obtain meaningful positive results (Figure 2A).
In order to further study and analyze the effect of DLGAP5 expression on prognosis, the data from the TCGA database were used to analyze the effect of DLGAP5 on overall survival and Progression-Free Survival in glioblastomas and low-grade glioma. The data showed that in glioblastoma and low-grade glioma, the overall survival and Progression-Free Survival of the DLGAP5 high-expression group was significantly lower than that of the low-expression group (p < 0.001), (Figure 2B). We extracted relevant data from the TCGA database to analyze the expression difference of DLGAP5 in different grades of glioma and its relationship with the prognosis of glioma patients. It can be seen from Figure 2C that the high expression of DLGAP5 in WHO grade 3 glioma is higher than that in WHO grade 2 glioma, the death rate of the DLGAP5 high expression group is higher than that of the low expression group, and the survival rate of low expression group is significantly higher than DLGAP5 high expression group.
It is known from this picture that DLGAP5 has an adverse effect on the prognosis of patients with glioma. It is further verified that DLGAP5 is an oncogene in glioma.
Genetic variation of DLGAP5 in glioma
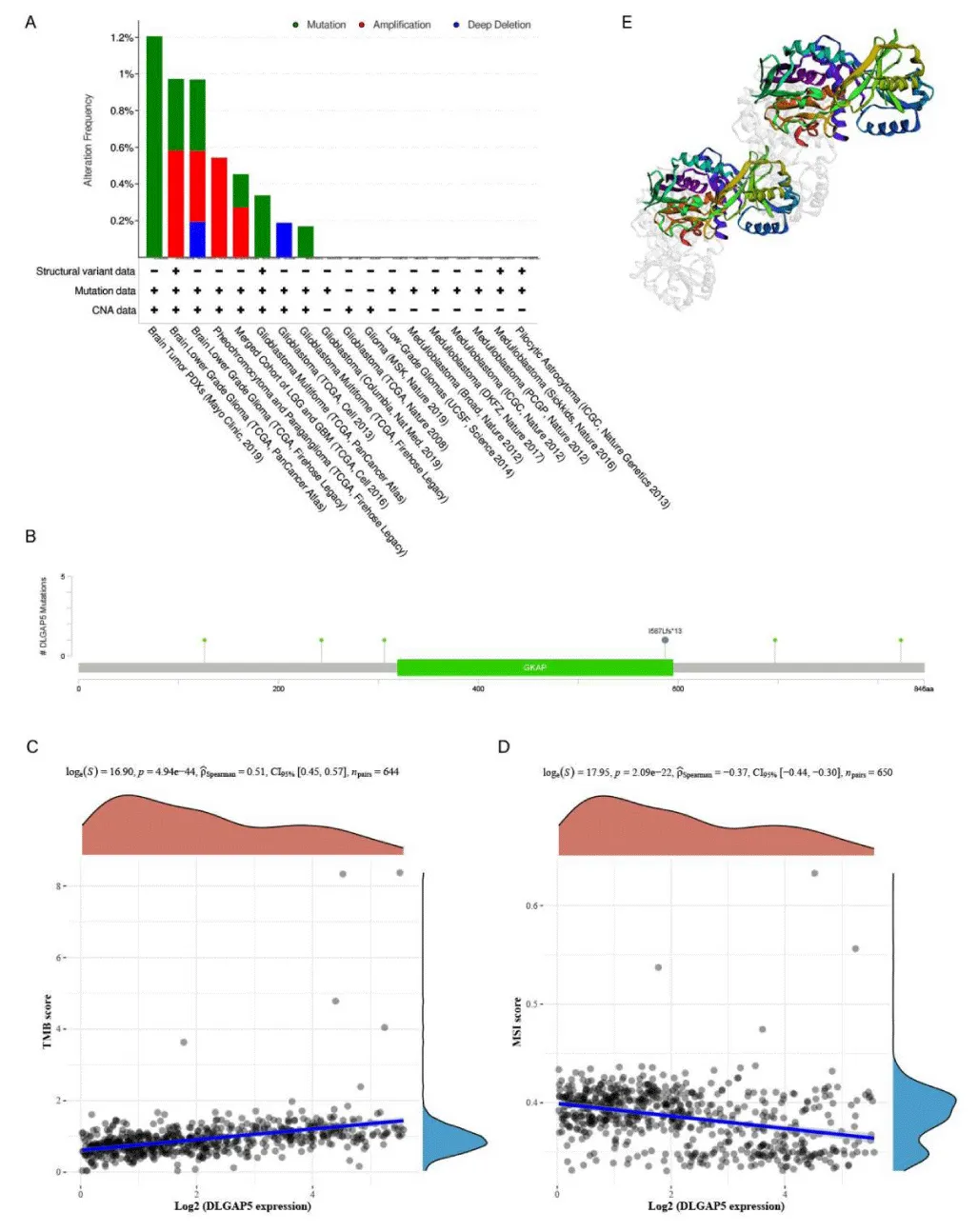
In this study, the cBioPortal database was used to study the variation of the DLGAP5 gene in glioma. DLGAP5 changes in glioma were studied in detail by analyzing 6,339 patients and 6,548 samples from 20 studies in the database. As shown in the Figure 3A histogram, the horizontal axis represents 18 valuable studies filtered out of 20 studies, and the vertical axis is the mutation frequency. The three colors in the picture represent different types of variation. According to the 2019 Mayo Clinic’s PDXs study of brain tumors, 1.2% of the DLGAP5 gene mutations were found. In low-grade brain glioma, TCGA tumor data showed that DLGAP5 gene amplification accounted for 0.58%, and mutation accounted for 0.39%. According to these studies, it can be seen that the main variations of the DLGAP5 gene in glioma are gene mutation, gene amplification, and gene deep deletion. Then, we used cBioPortal database to research the mutation sites of the DLGAP5 gene and which amino acids changed. As shown in Figure 3C, there are a total of 10 mutations, including 4 repeated mutations in patients with multiple samples. Among the 10 variants, 9 were missense, and 1 was truncation. This figure shows that 6 variations sited in the DLGAP5 gene, which lead to the changes of amino acids of D126H, P243S, S306F, I1587Lfs*13, P697T, and Q823L, in which the variation site of I1587Lfs*13 is located at the site of guanosine monophosphate kinase-associated protein (GKAP). Finally, it is displayed by 3D structure.
In this study, we analyzed the relationship between the expression of DLGAP5 and Tumor Mutation Burden and Microsatellite Instability and then explored the therapeutic effect of immunotherapy on gliomas expressing DLGAP5. As shown in the figure, the Abscissa represents the DLGAP5 expression distribution, the ordinate represents the TMB/MSI score distribution, the right density curve represents the TMB/MSI score distribution trend, and the upper-density curve represents the DLGAP5 expression distribution curve. The TMB score increased with the increase of DLGAP5 expression. It meant that the expression of DLGAP5 in glioma was positively correlated with TMB (p < 0.0001). However, the score of TMB is between 0 and 2mutations/Mb, and the TMB score of root drama is low. We can see from Figure 3D that the score of MSI decreased with the increase of DLGAP5 expression (p < 0.0001). However, the score of MSI ranged from 0 to 0.4. It was classified as MSI low-frequency type according to MSI classification.
Analysis of immune infiltration results
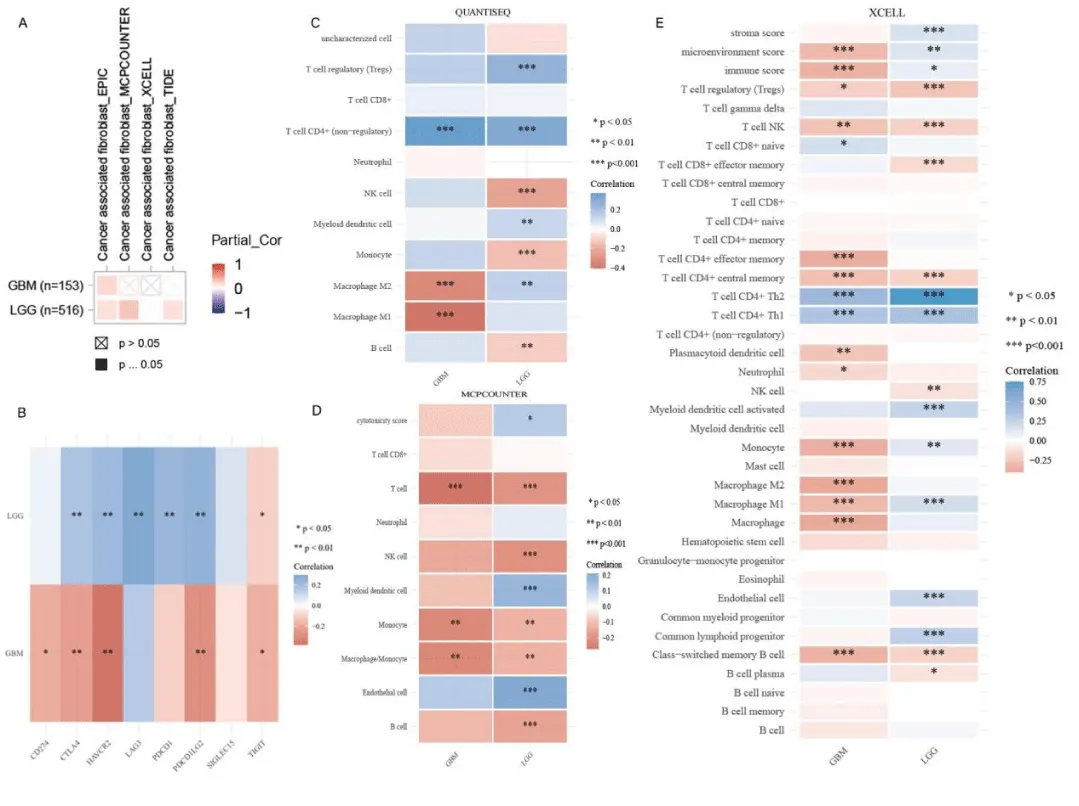
The growth and development of glioma are closely related to the tumor microenvironment. The infiltration level of tumor-associated fibroblasts in DLGAP5-expressing gliomas was investigated by using different algorithms in TIMER2.0. The different colors in the picture represented different relationships. The reddish color indicated a positive correlation and the blue color indicated a negative correlation. In glioblastoma, only the EPIC algorithm was positively correlated, while there was no statistically significant association in the other three algorithms. In low-grade glioma, three algorithms, EPIC, MCPCOUNTER, and TIDE, indicated that DLGAP5-expressing low-grade gliomas positively correlated with tumor-associated fibroblasts, (Figure 4A). In Figure 4B, we investigated the link between gliomas and immune checkpoints. SIGLEC15, TIGIT, CD274, HAVCR2, PDCD1, CTLA4, LAG3, and PDCD1LG2 were genes involved in immune checkpoints. In Figure 4B, the blue color indicated a positive correlation, and the red color indicated a negative correlation. In glioblastoma, there was no statistical association with LAG3, PDCD1, and SIGLEC15, but negative correlation with other immune checkpoint genes and a significant negative correlation with HAVCR2 (P<0.001). In low-grade glioma, LGG was negatively correlated with TIGIT and had no statistical significance with CD274 and SIGLEC15. But it was positively correlated with other genes, and significantly positively correlated with LAG3 (P<0.01). In this study, the level of immune cell infiltration in glioma was also investigated by using different algorithms. In the QUANTISEQ, MCPCOUNTER, and XCELL algorithms, the blue color indicated a positive correlation and the red color indicated a negative correlation. Among the three algorithms with statistical significance, most immune cell infiltration levels in glioblastoma were negatively correlated, especially the macrophage infiltration level in glioblastoma was significantly negatively correlated (p < 0.01). In low-grade gliomas, there were multiple positive correlations of immune cell infiltration and negative correlations of immune cell infiltration, (Figure 4CDE).
Functional analysis
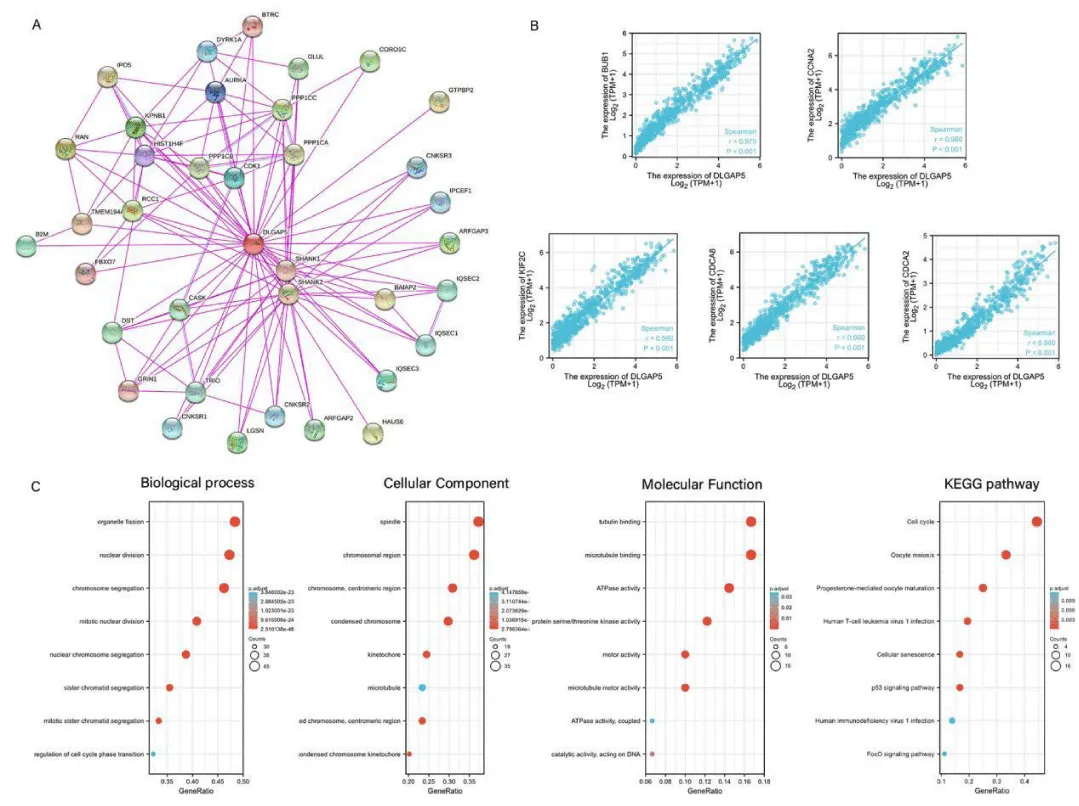
In this study, the DLGAP5 protein interaction network was studied by using the String database. There was a total of 37 protein nodes as shown in Figure 5A. Each node represented all proteins encoded by a single protein-coding locus. The red node in the middle was the protein encoded by the DLGAP5-encoding gene. The lines between the nodes represented protein-protein associations, indicating that they collectively participated in a certain function. The color of the line indicated the source of evidence for the association between proteins, and pink indicated experimental validation. The blue indicated evidence from the database. The figure showed that DLGAP5 had experimentally verified associations with other 36 nodes. The top 100 genes related to the expression of the DLGAP5 gene were obtained from the glioma data of the TCGA database. We selected the top five genes of BUB1, CCNA2, KIF2C, CDCA8, and CDCA2. As shown in Figure 5B, the abscissa in the figure represented the expression level of DLGAP5, and the ordinate represents the expression level of the top 5 related genes. With the increase of DLGAP5 gene expression, the expression of the top 5 genes related to its expression also increased. Therefore, we thought BUB1, CCNA2, KIF2C, CDCA8, CDCA2 were positively correlated with DLGAP5 (p < 0.001).
The gene ultimately exerted its final function by affecting the encoded protein. Therefore, we performed DLGAP5 gene enrichment analysis to research the DLGAP5 expression information in this study. In the DLGAP5 gene enrichment analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis and Gene Ontology (GO) enrichment analysis were mainly performed. The Gene Ontology enrichment contained cellular components, molecular functions, and biological processes. The abscissa in the figure was the Gene Ratio and the ordinate was the enrichment analysis item in the figure. In Figure 5C, the color of the circle represented the correlation. The reddish color indicated a higher correlation. In terms of molecular function, DLGAP5 was mainly related to tubulin binding, microtubule binding, and adenosine triphosphatase activity. In the biological process, the gene is mainly associated with the organelle fission process, nuclear division process, and chromosome segregation process. In cellular components, the gene was mainly associated with spindles, chromosomal regions, and centromeric regions. In the Encyclopedia of Genomes enrichment analysis, DLGAP5 is mainly associated with cell cycle, oocyte meiosis, p53 signaling pathway, and progesterone-mediated oocyte maturation.
Discussion
Glioma is the most common type of primary central nervous system malignant tumor, which mainly originates from astrocytes, oligodendrocytes, and so on [33]. Due to the characteristics of rapid growth and aggressive glioma, glioma patients have poorer survival time and quality of life compared with other nervous system tumors. The median survival for glioblastoma is only 14.4 months [34]. The standard treatment for gliomas is maximal resection, followed by standard radiotherapy combined with temozolomide chemotherapy. Despite standard treatment, the survival and quality of life of glioma patients have not improved significantly. With the development of basic medical research and clinical science, new therapeutic schemes have been applied in the adjuvant treatment of gliomas, such as immunotherapy, targeted therapy, and tumor treatment fields. These cutting-edge adjuvant therapies have prolonged survival and improved the quality of life of patients with glioma [33]. At present, molecular targeted therapy for glioma has entered the stage of clinical trials, but few positive effects [35]. Therefore, finding suitable target genes has positive significance for the treatment of glioma. A host of researchers have found that DLGAP5 plays an important role in the occurrence and development of other tumors such as lung cancer, cervical cancer, and intestinal cancer. In this study, the effects of DLGAP5 on the occurrence and progression of glioma were analyzed from the aspects of DLGAP5 gene expression, prognosis, immune response, and mechanism of action.
DLGAP5, also known as DLG7 or HURP, is a mitotic spindle protein whose function is to promote the formation of tubulin polymers [7]. As a cell cycle-regulating gene, DLGAP5 mRNA changes periodically in the cell cycle and plays an important role in the occurrence and development of a variety of tumors. In hepatocellular carcinoma, researchers found that DLGAP5 was overexpressed in hepatocellular carcinoma, and silencing of DLGAP5 inhibited the HCC cycle and proliferation of hepatocellular carcinoma differentiation [15]. By flow cytometry, some research teams found that after DLGAP5 down-regulation in glioma, the cells in G0/G1 phase increased, while the cells in S and G2/M phases decreased significantly. Their study suggested that DLGAP5 may contribute to glioma progression by accelerating G0/G1 phase progression [17]. However, the specific mechanism of DLGAP5 in glioma remains unclear. In this study, we mainly explored the possible molecular mechanism of DLGAP5.
Through the analysis of TCGA and CGGA databases, it was found that DLGAP5 was highly expressed in glioma tissues and lowly expressed in normal tissues. We found that the expression of DLGAP5 in tumor tissues increased with the increase of glioma grade. The differential expression of DLGAP5 in tumor tissues and normal neural tissues suggested that DLGAP5 may be a gene that promotes glioma development. In the prognostic analysis, the Overall Survival and Progression Free Survival of glioma patients in the high-expression group of DLGAP5 were lower than those in the low-expression group. In order to further explore the mechanism of DLGAP5 in glioma tissue, we carried out a detailed exploration of different aspects of DLGAP5 gene mutation, immune cell infiltration analysis, gene function analysis, and so on. The occurrence of tumors is caused by related gene mutations, and in this process, tumor suppressor genes and tumor-promoting genes are mainly involved. The mutation of DLGAP5 changed the downstream proteins, causing the occurrence and progression of glioma. By analyzing the data in the TCGA database, the main types of DLGAP5 gene variants were gene mutation, gene amplification, and gene deep deletion. The mutated genes exert corresponding functions through transcription and translation into proteins. In this study, we confirmed the major mutation sites of DLGAP5. Epigenetics is the process of causing heritable gene expression or cell phenotype changes through some mechanism without changing the DNA sequence, which includes DNA methylation, histone modification, and microRNA [36]. Epigenetic alterations are the main mechanisms underlying many human diseases, especially growth and developmental disorders, including Beckwith-Wiedemann (BWS), Silver-Russell, Prader-Willi, and Angelman syndromes [37]. The role of epigenetic changes in DLGAP5 variation in glioma needs to be further explored in basic experiments.
At present, studies have confirmed that Tumor Mutation Burden (TMB), Microsatellite Instability (MSI), and Tumor Micro-Environment are independent prognostic indicators to evaluate the prognosis of patients and the effect of immunotherapy. TMB refers to the number of genetic mutations in tumor cells. It represents the number of somatic nonsynonymous mutations in a particular genomic region, usually expressed in mutations per megabase (mut/Mb). The tumor mutation load can laterally reflect the tumor’s ability to generate neoantigens and can be used to predict the immunotherapy effect of various tumors. In general, the higher the tumor mutational burden, the better the effect of immunotherapy [38]. MSI refers to the change of any length of microsatellites caused by the insertion or deletion of base pairs in the microsatellite region due to point mutation, sliding chain mismatch, and mismatch repair mechanisms during DNA replication in tumor tissues. According to the level of MSI, MSI can be divided into three types: low microsatellite instability (MSI-L), high microsatellite instability (MSI-H), and microsatellite stability (MSS). Tumors with high microsatellite instability are more effective in immunotherapy than those with low microsatellite instability [39,40]. TMB and MSI are commonly used as biomarkers of immunotherapy to predict the therapeutic effect of immunotherapy on tumors. In DLGAP5-expressing glioma, the number of TMB was lower, and the level of MSI was low microsatellite instability. TME refers to the surrounding microenvironment of tumor cell growth and development, including fibroblasts, immune cells, surrounding blood vessels, and tumor cells. The tumor microenvironment plays an important role in tumor growth and development and the body’s response to tumor cells [41].
Tumor-Associated Fibroblasts (CAFs) are a core component of the tumor microenvironment. CAFs play an important role in tumor growth and development. The main role of CAFs is to promote tumor growth, however, under certain circumstances, they can have tumor-suppressive effects [42,43]. This study analyzed the infiltration levels of tumor-associated fibroblasts in the glioblastoma and low-grade glioma microenvironments. CAFs were positively correlated with DLGAP5 expression in both glioblastoma and low-grade glioma. As an important part of the body, immune cells play an important role in killing tumor cells in the body. Immune Checkpoints (ICP) refer to a series of molecules expressed on immune cells that can regulate the degree of immune activation. ICP plays an important role in maintaining the body’s self-tolerance, preventing immune responses, and controlling the timing and extent of immune responses. ICP is expressed on immune cells and inhibits the function of immune cells by interfering with costimulatory signals so that the body cannot produce an effective anti-tumor immune response, and tumors thus form immune escape [44]. It is a new treatment for tumors to block the inhibitory effect of tumor cells on the immune system by inhibiting immune checkpoints.
By studying the expression levels of immune checkpoint-related genes in glioma, we analyzed the immune response of the body to glioma patients. In glioblastoma, most of the statistically significant immune checkpoints were negatively expressed. In low-grade gliomas, the majority were positive. Inhibition of immune checkpoints with immune checkpoint inhibitors may have dramatic effects in low-grade gliomas. Whereas in glioblastoma, the effects may be minimal.
Protein-protein interaction network (PPI) is composed of proteins interacting with each other to participate in all aspects of life processes such as biological signal transmission, regulation of gene expression, metabolism of substances and energy, and regulation of cell cycle. PPI is of great significance to people’s understanding of protein function and relationship [45]. By analyzing the interaction between proteins in glioma, it is of great significance to understand the function of proteins in glioma and the relationship between proteins. The String database showed that the DLGAP5-expressed protein is closely linked to 36 other proteins in glioma. Kyoto Encyclopedia of Genes and Genomes (KEGG) is a database for systematic analysis of gene function and genomic information, which integrates information from genomics, biochemistry, etc. It is important to study the process of gene and expression information as a whole [46]. Normal human cells undergo normal cell cycles after receiving appropriate mitotic signals. In the case of tumor cells, the normal cell cycle fails to proceed to lead to tumorigenesis [47]. As a transcription factor composed of different domains, P53 plays a central role in maintaining cellular homeostasis and is often deregulated in cancer [48]. Studies have confirmed that the p53 pathway is deregulated in most glioblastoma [49]. The p53 pathway plays an important role in the occurrence and progression of glioma. Exploring the molecular mechanism of DLGAP5 is of great significance for understanding the relationship between DLGAP5 and glioma. Gene Ontology (GO) covers the molecular functions, cellular components, and biological functions of genes. Molecular Function (MF) is to describe the molecular biological function of the body; Cell Component (CC) refers to the functional part of the downstream substance of the gene; Biological Process (BP) is the process analysis of molecular function from generation to function [50]. We found that the main mechanism of DLGAP5 in Biological Processes is involved in organelle fission, nuclear division, and chromosome segregation. In terms of Molecular Function, DLGAP5 is mainly involved in tubulin binding, microtubule binding, ATPase activity, and microtubule motor activity. In terms of the Cellular Component, DLGAP5 is involved in the spindle, chromosomal region, and condensed chromosome. KEGG analysis showed that DLGAP5 regulated glioma progression by participating in the cell cycle, oocyte meiosis, p53 signaling pathway, and so on. A large number of studies have confirmed that the genetic variation of glioma is finally pooled in three core cell signal transduction pathways: RTK/RAS/PI-3K pathway, p53 pathway, and RB pathway [51]. Therefore, DLGAP5 may regulate the occurrence and progression of glioma through the p53 pathway and cell cycle.
This study first analyzed the expression of DLGAP5 in glioma tissues and normal neural tissues. And then we explored the relationship between the expression level of DLGAP5 and the prognosis of glioma patients. Through the analysis of gene mutation, immune cell infiltration, and functional enrichment of DLGAP5, it was finally concluded that DLGAP5 may regulate the progression of glioma through the p53 pathway and cell cycle. Although relevant studies have proved that DLGAP5 plays an important role in the development of glioma, the specific mechanism of action is still unclear. In this study, the molecular mechanism of DLGAP5 was studied for the first time, and the results of further study were obtained. A number of basic experiments have confirmed the role of this gene in many tumors, including glioma. However, the mechanism of this gene in glioma remains unclear. Our study was conducted on the basis of multiple databases and online analysis sites. Although it lacks the research confirmation of basic experiments, it can be used as a supplement to other studies. Therefore, we need to further validate our results through basic experiments, so as to provide a new scheme for the clinical treatment of glioma.
Conclusion
Our findings indicate that DLGAP5 is highly expressed in glioma tissues. Moreover, the expression level of DLGAP5 increases with the grade of glioma. The high expression of DLGAP5 is closely related to the low prognosis of glioma patients. DLGAP5 may act on glioma through the cell cycle and p53 pathway.
Kangjie Du, Yu Zhang, and Mengyao contributed equally to this study. All authors read and approved the final manuscript.
- Ostrom QT, Bauchet L, Davis FG, Deltour I, Fisher JL, Langer CE, Pekmezci M, Schwartzbaum JA, Turner MC, Walsh KM, Wrensch MR, Barnholtz-Sloan JS. The epidemiology of glioma in adults: a "state of the science" review. Neuro Oncol. 2014 Jul;16(7):896-913. doi: 10.1093/neuonc/nou087. PMID: 24842956; PMCID: PMC4057143.
- Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM, Reifenberger G, Soffietti R, von Deimling A, Ellison DW. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol. 2021 Aug 2;23(8):1231-1251. doi: 10.1093/neuonc/noab106. PMID: 34185076; PMCID: PMC8328013.
- Bush NA, Chang SM, Berger MS. Current and future strategies for treatment of glioma. Neurosurg Rev. 2017 Jan;40(1):1-14. doi: 10.1007/s10143-016-0709-8. Epub 2016 Apr 16. PMID: 27085859.
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005 Mar 10;352(10):987-96. doi: 10.1056/NEJMoa043330. PMID: 15758009.
- Aldape K, Zadeh G, Mansouri S, Reifenberger G, von Deimling A. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015 Jun;129(6):829-48. doi: 10.1007/s00401-015-1432-1. Epub 2015 May 6. PMID: 25943888.
- Bassal S, Nomura N, Venter D, Brand K, McKay MJ, van der Spek PJ. Characterization of a novel human cell-cycle-regulated homologue of Drosophila dlg1. Genomics. 2001 Sep;77(1-2):5-7. doi: 10.1006/geno.2001.6570. PMID: 11543626.
- Santarella RA, Koffa MD, Tittmann P, Gross H, Hoenger A. HURP wraps microtubule ends with an additional tubulin sheet that has a novel conformation of tubulin. J Mol Biol. 2007 Feb 2;365(5):1587-95. doi: 10.1016/j.jmb.2006.10.064. Epub 2006 Oct 25. PMID: 17118403.
- Wong J, Fang G. HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J Cell Biol. 2006 Jun 19;173(6):879-91. doi: 10.1083/jcb.200511132. Epub 2006 Jun 12. PMID: 16769820; PMCID: PMC2063914.
- Ye F, Tan L, Yang Q, Xia Y, Deng LW, Murata-Hori M, Liou YC. HURP regulates chromosome congression by modulating kinesin Kif18A function. Curr Biol. 2011 Sep 27;21(18):1584-91. doi: 10.1016/j.cub.2011.08.024. Epub 2011 Sep 14. PMID: 21924616.
- Wilde A. "HURP on" we're off to the kinetochore! J Cell Biol. 2006 Jun 19;173(6):829-31. doi: 10.1083/jcb.200605150. PMID: 16785318; PMCID: PMC2063907.
- Marumoto T, Zhang D, Saya H. Aurora-A - a guardian of poles. Nat Rev Cancer. 2005 Jan;5(1):42-50. doi: 10.1038/nrc1526. PMID: 15630414.
- Wong J, Lerrigo R, Jang CY, Fang G. Aurora A regulates the activity of HURP by controlling the accessibility of its microtubule-binding domain. Mol Biol Cell. 2008 May;19(5):2083-91. doi: 10.1091/mbc.e07-10-1088. Epub 2008 Mar 5. PMID: 18321990; PMCID: PMC2366856.
- Branchi V, García SA, Radhakrishnan P, Győrffy B, Hissa B, Schneider M, Reißfelder C, Schölch S. Prognostic value of DLGAP5 in colorectal cancer. Int J Colorectal Dis. 2019 Aug;34(8):1455-1465. doi: 10.1007/s00384-019-03339-6. Epub 2019 Jul 8. PMID: 31286215.
- Tagal V, Wei S, Zhang W, Brekken RA, Posner BA, Peyton M, Girard L, Hwang T, Wheeler DA, Minna JD, White MA, Gazdar AF, Roth MG. SMARCA4-inactivating mutations increase sensitivity to Aurora kinase A inhibitor VX-680 in non-small cell lung cancers. Nat Commun. 2017 Jan 19;8:14098. doi: 10.1038/ncomms14098. PMID: 28102363; PMCID: PMC5253647.
- Breuer M, Kolano A, Kwon M, Li CC, Tsai TF, Pellman D, Brunet S, Verlhac MH. HURP permits MTOC sorting for robust meiotic spindle bipolarity, similar to extra centrosome clustering in cancer cells. J Cell Biol. 2010 Dec 27;191(7):1251-60. doi: 10.1083/jcb.201005065. Epub 2010 Dec 20. PMID: 21173113; PMCID: PMC3010075.
- Zheng R, Shi Z, Li W, Yu J, Wang Y, Zhou Q. Identification and prognostic value of DLGAP5 in endometrial cancer. PeerJ. 2020 Nov 27;8:e10433. doi: 10.7717/peerj.10433. PMID: 33312770; PMCID: PMC7703392.
- Zhou D, Wang M, Zhang Y, Wang K, Zhao M, Wang Y, Wang X, Yu R, Zhou X. Screening and identification of LMNB1 and DLGAP5, two key biomarkers in gliomas. Biosci Rep. 2021 May 28;41(5):BSR20210231. doi: 10.1042/BSR20210231. PMID: 33956061; PMCID: PMC8144940.
- Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020 Jul 2;48(W1):W509-W514. doi: 10.1093/nar/gkaa407. PMID: 32442275; PMCID: PMC7319575.
- Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017 Nov 1;77(21):e108-e110. doi: 10.1158/0008-5472.CAN-17-0307. PMID: 29092952; PMCID: PMC6042652.
- Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, Jiang P, Shen H, Aster JC, Rodig S, Signoretti S, Liu JS, Liu XS. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016 Aug 22;17(1):174. doi: 10.1186/s13059-016-1028-7. PMID: 27549193; PMCID: PMC4993001.
- Zhao Z, Zhang KN, Wang Q, Li G, Zeng F, Zhang Y, Wu F, Chai R, Wang Z, Zhang C, Zhang W, Bao Z, Jiang T. Chinese Glioma Genome Atlas (CGGA): A Comprehensive Resource with Functional Genomic Data from Chinese Glioma Patients. Genomics Proteomics Bioinformatics. 2021 Feb;19(1):1-12. doi: 10.1016/j.gpb.2020.10.005. Epub 2021 Mar 2. PMID: 33662628; PMCID: PMC8498921.
- Bao ZS, Chen HM, Yang MY, Zhang CB, Yu K, Ye WL, Hu BQ, Yan W, Zhang W, Akers J, Ramakrishnan V, Li J, Carter B, Liu YW, Hu HM, Wang Z, Li MY, Yao K, Qiu XG, Kang CS, You YP, Fan XL, Song WS, Li RQ, Su XD, Chen CC, Jiang T. RNA-seq of 272 gliomas revealed a novel, recurrent PTPRZ1-MET fusion transcript in secondary glioblastomas. Genome Res. 2014 Nov;24(11):1765-73. doi: 10.1101/gr.165126.113. Epub 2014 Aug 18. PMID: 25135958; PMCID: PMC4216918.
- Zhao Z, Meng F, Wang W, Wang Z, Zhang C, Jiang T. Comprehensive RNA-seq transcriptomic profiling in the malignant progression of gliomas. Sci Data. 2017 Mar 14;4:170024. doi: 10.1038/sdata.2017.24. PMID: 28291232; PMCID: PMC5349247.
- Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat Med. 2011 Mar;17(3):297-303. doi: 10.1038/nm.2323. PMID: 21383744.
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013 Apr 2;6(269):pl1. doi: 10.1126/scisignal.2004088. PMID: 23550210; PMCID: PMC4160307.
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012 May;2(5):401-4. doi: 10.1158/2159-8290.CD-12-0095. Erratum in: Cancer Discov. 2012 Oct;2(10):960. PMID: 22588877; PMCID: PMC3956037.
- Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000 Sep 15;28(18):3442-4. doi: 10.1093/nar/28.18.3442. PMID: 10982861; PMCID: PMC110752.
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013 Jan;41(Database issue):D808-15. doi: 10.1093/nar/gks1094. Epub 2012 Nov 29. PMID: 23203871; PMCID: PMC3531103.
- Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, Jensen LJ, von Mering C. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021 Jan 8;49(D1):D605-D612. doi: 10.1093/nar/gkaa1074. Erratum in: Nucleic Acids Res. 2021 Oct 11;49(18):10800. PMID: 33237311; PMCID: PMC7779004.
- Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019 Jul 2;47(W1):W556-W560. doi: 10.1093/nar/gkz430. PMID: 31114875; PMCID: PMC6602440.
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. Epub 2003 Apr 3. PMID: 12734009.
- Zhang RJ, Li Y, Liu Q, Gao YJ, Du J, Ma J, Sun SG, Wang L. Differential Expression Profiles and Functional Prediction of Circular RNAs and Long Non-coding RNAs in the Hippocampus of Nrf2-Knockout Mice. Front Mol Neurosci. 2019 Aug 9;12:196. doi: 10.3389/fnmol.2019.00196. PMID: 31447646; PMCID: PMC6697070.
- Jiang T, Nam DH, Han L, Wang Q. On behalf of the Chinese Glioma Cooperative Group (CGCG), Society for Neuro-Oncology of China (SNO-China), Chinese Brain Cancer Association (CBCA), Chinese Glioma Genome Atlas (CGGA), Asian Glioma Genome Atlas (AGGA) network, Clinical practice guidelines for the management of adult diffuse gliomas, Cancer Letters 2020.
- Yang P, Wang Y, Peng X, You G, Zhang W, Yan W, Bao Z, Wang Y, Qiu X, Jiang T. Management and survival rates in patients with glioma in China (2004-2010): a retrospective study from a single-institution. J Neurooncol. 2013 Jun;113(2):259-66. doi: 10.1007/s11060-013-1103-9. Epub 2013 Mar 13. PMID: 23483435.
- Nakada M, Kita D, Teng L, Pyko IV, Watanabe T, Hayashi Y, Hamada JI. Receptor Tyrosine Kinases: Principles and Functions in Glioma Invasion. Adv Exp Med Biol. 2020;1202:151-178. doi: 10.1007/978-3-030-30651-9_8. PMID: 32034713.
- Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006 Jan;7(1):21-33. doi: 10.1038/nrg1748. PMID: 16369569.
- Miranda Furtado CL, Dos Santos Luciano MC, Silva Santos RD, Furtado GP, Moraes MO, Pessoa C. Epidrugs: targeting epigenetic marks in cancer treatment. Epigenetics. 2019 Dec;14(12):1164-1176. doi: 10.1080/15592294.2019.1640546. Epub 2019 Jul 13. PMID: 31282279; PMCID: PMC6791710.
- Ritterhouse LL. Tumor mutational burden. Cancer Cytopathol. 2019 Dec;127(12):735-736. doi: 10.1002/cncy.22174. Epub 2019 Aug 21. PMID: 31433548.
- Yang G, Zheng RY, Jin ZS. Correlations between microsatellite instability and the biological behaviour of tumours. J Cancer Res Clin Oncol. 2019 Dec;145(12):2891-2899. doi: 10.1007/s00432-019-03053-4. Epub 2019 Oct 15. PMID: 31617076; PMCID: PMC6861542.
- Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998 Nov 15;58(22):5248-57. PMID: 9823339.
- Garufi A, Traversi G, Cirone M, D'Orazi G. HIPK2 role in the tumor-host interaction: Impact on fibroblasts transdifferentiation CAF-like. IUBMB Life. 2019 Dec;71(12):2055-2061. doi: 10.1002/iub.2144. Epub 2019 Aug 15. PMID: 31414572; PMCID: PMC6899452.
- Lin Y, Cai Q, Chen Y, Shi T, Liu W, Mao L, Deng B, Ying Z, Gao Y, Luo H, Yang X, Huang X, Shi Y, He R. CAFs shape myeloid-derived suppressor cells to promote stemness of intrahepatic cholangiocarcinoma through 5-lipoxygenase. Hepatology. 2022 Jan;75(1):28-42. doi: 10.1002/hep.32099. Epub 2021 Dec 5. PMID: 34387870.
- Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM, Fearon D, Greten FR, Hingorani SR, Hunter T, Hynes RO, Jain RK, Janowitz T, Jorgensen C, Kimmelman AC, Kolonin MG, Maki RG, Powers RS, Puré E, Ramirez DC, Scherz-Shouval R, Sherman MH, Stewart S, Tlsty TD, Tuveson DA, Watt FM, Weaver V, Weeraratna AT, Werb Z. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer. 2020 Mar;20(3):174-186. doi: 10.1038/s41568-019-0238-1. Epub 2020 Jan 24. PMID: 31980749; PMCID: PMC7046529.
- Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018 Mar 23;359(6382):1350-1355.
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019 Jan 8;47(D1):D607-D613. doi: 10.1093/nar/gky1131. PMID: 30476243; PMCID: PMC6323986.
- Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019 Jan 8;47(D1):D590-D595. doi: 10.1093/nar/gky962. PMID: 30321428; PMCID: PMC6324070.
- Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001 May 17;411(6835):342-8. doi: 10.1038/35077213. PMID: 11357141.
- Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014 May;14(5):359-70. doi: 10.1038/nrc3711. Epub 2014 Apr 17. PMID: 24739573; PMCID: PMC4049238.
- Zhang Y, Dube C, Gibert M Jr, Cruickshanks N, Wang B, et al. The p53 Pathway in Glioblastoma. Cancers (Basel). 2018 Sep 1;10(9):297.
- Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019 Jan 8;47(D1):D419-D426. doi: 10.1093/nar/gky1038. PMID: 30407594; PMCID: PMC6323939.
- Wang LB, Karpova A, Gritsenko MA, Kyle JE, Cao S, Li Y, Rykunov D, Colaprico A, Rothstein JH, Hong R, Stathias V, Cornwell M, Petralia F, Wu Y, Reva B, Krug K, Pugliese P, Kawaler E, Olsen LK, Liang WW, Song X, Dou Y, Wendl MC, Caravan W, Liu W, Cui Zhou D, Ji J, Tsai CF, Petyuk VA, Moon J, Ma W, Chu RK, Weitz KK, Moore RJ, Monroe ME, Zhao R, Yang X, Yoo S, Krek A, Demopoulos A, Zhu H, Wyczalkowski MA, McMichael JF, Henderson BL, Lindgren CM, Boekweg H, Lu S, Baral J, Yao L, Stratton KG, Bramer LM, Zink E, Couvillion SP, Bloodsworth KJ, Satpathy S, Sieh W, Boca SM, Schürer S, Chen F, Wiznerowicz M, Ketchum KA, Boja ES, Kinsinger CR, Robles AI, Hiltke T, Thiagarajan M, Nesvizhskii AI, Zhang B, Mani DR, Ceccarelli M, Chen XS, Cottingham SL, Li QK, Kim AH, Fenyö D, Ruggles KV, Rodriguez H, Mesri M, Payne SH, Resnick AC, Wang P, Smith RD, Iavarone A, Chheda MG, Barnholtz-Sloan JS, Rodland KD, Liu T, Ding L; Clinical Proteomic Tumor Analysis Consortium. Proteogenomic and metabolomic characterization of human glioblastoma. Cancer Cell. 2021 Apr 12;39(4):509-528.e20. doi: 10.1016/j.ccell.2021.01.006. Epub 2021 Feb 11. PMID: 33577785; PMCID: PMC8044053.
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
