Annals of Molecular and Genetic Medicine
Computational Analysis of Conserved Plant microRNA408 and Evaluation of its Cross-Kingdom Regulatory Potential in Humans
1MSc Bioinformatics, Stella Maris College, Chennai, India
2Department of Biotechnology, School of Life Sciences, Pondicherry University, Kalapet, Puducherry,India
Author and article information
Cite this as
Mitra S, Shukla LI. Computational Analysis of Conserved Plant microRNA408 and Evaluation of its Cross-Kingdom Regulatory Potential in Humans. Ann Mol Genet Med . 2025; 9(1): 001-015. Available from: 10.17352/amgm.000015
Copyright License
© 2025 Mitra S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Introduction: MicroRNA408 (miRNA408) is a highly conserved plant microRNA involved in copper homeostasis, development, and stress response. Given its presence in food crops and its evolutionary conservation, this study explores in silico analysis of mature and precursor miRNA408 (pre-miRNA408) and whether miRNA408 could exert post-transcriptional regulatory effects on human genes via dietary cross-kingdom interaction.
Methodology: A total of 72 mature and 55 precursor miRNA408 sequences were retrieved from miRBase v22. Sequence conservation, alignment, and phylogeny were analyzed using WebLogo, MAFFT, and iTOL, respectively. RNAfold was used to assess thermodynamic stability. A consensus miR408-3p sequence obtained from the literature was used to predict human gene targets via psRNATarget. Only targets predicted to be cleaved were selected for validation based on alignment, seed complementarity, and 3′compensatory sites pairing.
Results: The 3p strand exhibited higher nucleotide conservation than the 5p strand. Most mature sequences began with adenosine, suggesting AGO2–AGO9 loading. Longer precursors displayed lower minimum free energy (MFE), indicating higher structural stability. The high-confidence human target genes are involved in critical processes like gene regulation, cytoskeletal organization, neural development, and intracellular transport, highlighting the potential for dietary miRNAs to influence human biology.
Conclusion: miR408 has been reported in dietary plants such as tomato (Solanum lycopersicum). Multiple studies support the uptake and activity of dietary miRNAs in mammals. This study provides computational evidence for miR408’s cross-kingdom regulatory potential, which requires further experimental validation.
MicroRNAs (miRNAs) are small non-coding RNAs, typically 20–24 nucleotides in length, that play central roles in post-transcriptional gene regulation across eukaryotes [1-5]. They originate from precursor stem-loop structures that are processed by Dicer-like enzymes and subsequently incorporated into RNA-induced silencing complexes (RISCs). Within these complexes, miRNAs guide Argonaute (AGO) proteins to complementary mRNAs, resulting in either cleavage or translational repression depending on sequence complementarity [2,6]. In both plants and animals, the “seed region” spanning nucleotides 2–8 of the miRNA sequence is particularly important for target recognition [6-8].
In plants, miRNAs regulate diverse processes, including embryo patterning, organ development, responses to biotic and abiotic stresses, and nutrient homeostasis [2,9-11]. Conserved miRNAs, which are shared across multiple plant lineages, tend to have relatively high expression, low sequence variability, and essential developmental roles [12,13]. miR408 represents one of the most conserved plant miRNAs, reported in more than 30 species, and is strongly associated with copper homeostasis, stress tolerance, and growth regulation [12,10]. In Arabidopsis thaliana, the MIR408 gene is located on chromosome 2 (At2g47015), and its mature products, miR408-3p and miR408-5p, are processed from a conserved precursor hairpin [15].
Functional studies have identified key miR408 targets, including copper-binding proteins such as plastocyanin, uclacyanin, plantacyanin [1,9,12,13,14], and laccases [18]. Additional species-specific targets include TaTOC1s in wheat [13,15], DINUDT23 in longan [14,16], and IAA30 in rice [15,17]. By regulating these genes, miR408 has been shown to influence traits such as leaf size, petiole length, biomass, seed yield [12,16,18], heading time [13,15], root development [17,19], and somatic embryogenesis [14,16]. It also contributes to stress responses involving copper availability, light, cold, salinity, drought, osmotic stress, wounding, and nutrient limitation [12,19–22]. Overexpression of miR408 has been linked to enhanced stress tolerance and water-use efficiency [19,20,22]. Furthermore, recent work has revealed that the pri-miR408 transcript encodes miPEP408, a small peptide that enhances its own miRNA expression in a light-dependent manner through HY5, adding a regulatory dimension [29].
Bioinformatics approaches have greatly advanced the study of conserved plant miRNAs, enabling large-scale identification, expression profiling, structural analysis, and target prediction [31-33]. Several conserved families, including miR408, have been annotated and validated across crop and model plant species such as Triticum aestivum, Oryza sativa, Salvia miltiorrhiza, Populus trichocarpa, Arabidopsis thaliana, and Solanum lycopersicum using both computational pipelines and experimental assays [12,14-17,22,24]. For example, degradome sequencing in rice validated miR408-5p targets and demonstrated its role in flowering and stress responses [15,17], while studies in Populus highlighted its impact on biomass traits [12,14].
Beyond its roles in plants, recent findings point toward the possibility of dietary plant miRNAs participating in cross-kingdom regulation. Plant-derived miRNAs have been detected in mammalian serum and tissues following dietary intake, suggesting they can survive digestion and enter systemic circulation [35-38]. Zhang, et al. [34] reported that rice miR168a could target LDLRAP1 in mice, providing a proof of concept for interspecies regulation. Although subsequent studies have questioned the reproducibility of these findings [26-28], increasing evidence supports the potential of dietary plant miRNAs to influence animal or human gene expression.
Despite extensive functional characterization of miR408 in plants, its potential impact beyond the plant kingdom remains poorly understood. Considering its strong evolutionary conservation and structural stability, together with emerging evidence that plant miRNAs can persist in mammalian systems, miR408 represents a compelling candidate for cross-kingdom regulation. In this study, we employed a computational approach to evaluate the conservation and structural features of miR408 across multiple plant species, analyze its stability and AGO-binding potential, and predict possible human gene targets. These findings aim to provide new insights into the cross-kingdom regulatory potential of this conserved plant miRNA.
Materials and methodology
Retrieval of miR408 sequences
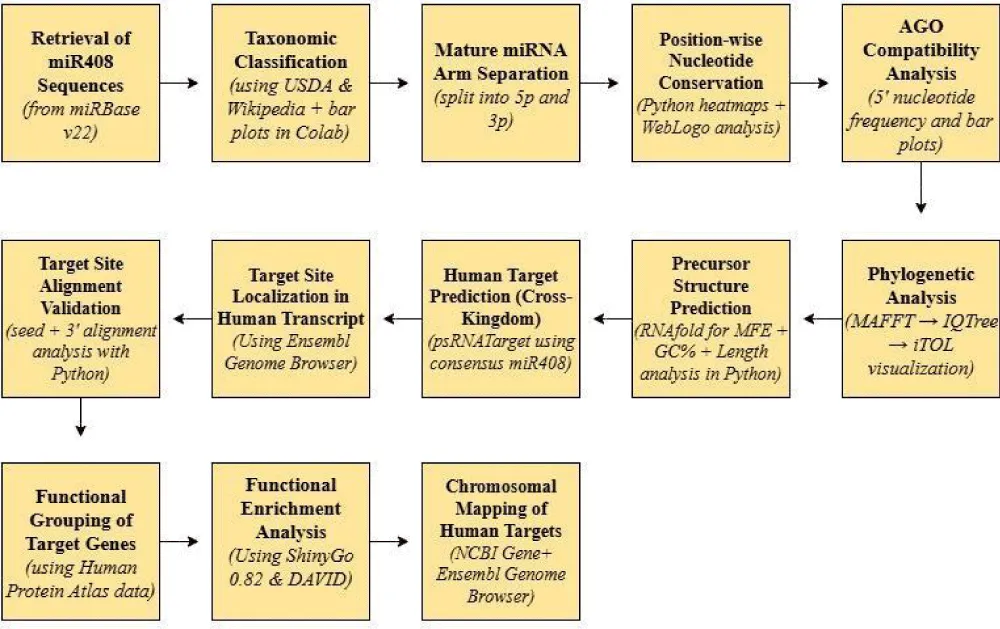
Mature and precursor sequences of miR408 were retrieved from the miRBase database (v22) (https://www.mirbase.org/) [32], a comprehensive repository of miRNA sequences and annotations from 271 organisms. A total of 73 mature and 55 precursor sequences were initially obtained, of which one mature entry was found to be obsolete and was removed. The final dataset comprised 72 mature and 55 precursor sequences, including 17 annotated as miR408-3p and miR408-5p, respectively. These sequences formed the basis for downstream analyses (Table 1, Figure 1). The overall pipeline was guided by established computational frameworks for miRNA analysis [41].
Taxonomic classification of source species
The species from which miR408 was annotated were classified taxonomically using the USDA Plants Database (https://www.aphis.usda.gov/). Taxonomic ranks, including division, class, order, and family, were assigned. Species richness across these ranks was visualized using bar and scatter plots generated in Google Colab with Matplotlib and Seaborn libraries. A complete dataset of taxonomic assignments is provided in Supplementary Table 1.
Position-specific nucleotide analysis
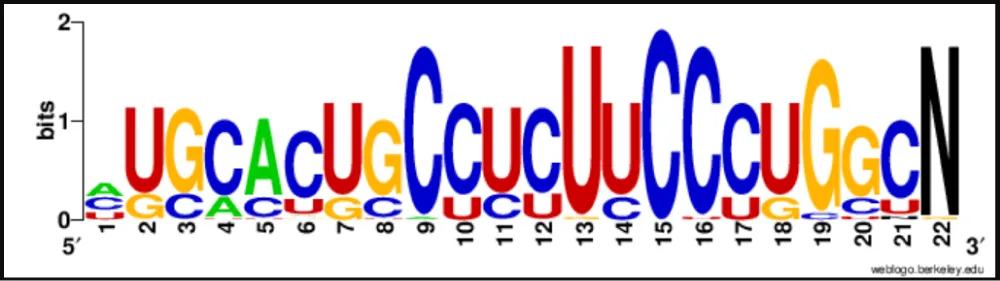
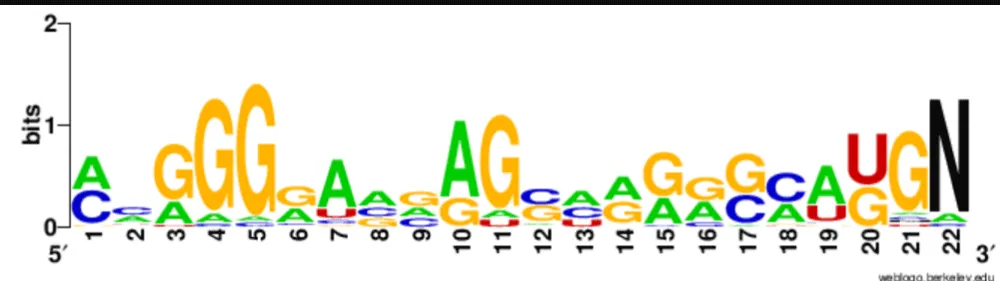
To assess sequence conservation, mature miR408 sequences were grouped into 5p and 3p arms based on their hairpin structures (Table 2). A custom Python script in Google Colab was used to analyze nucleotide frequencies across each position. Heatmaps were generated to depict position-wise conservation. WebLogo (https://weblogo.berkeley.edu/logo.cgi) [42] was further employed to visualize consensus sequence motifs, with “N” padding used to accommodate length variations.
AGO compatibility prediction
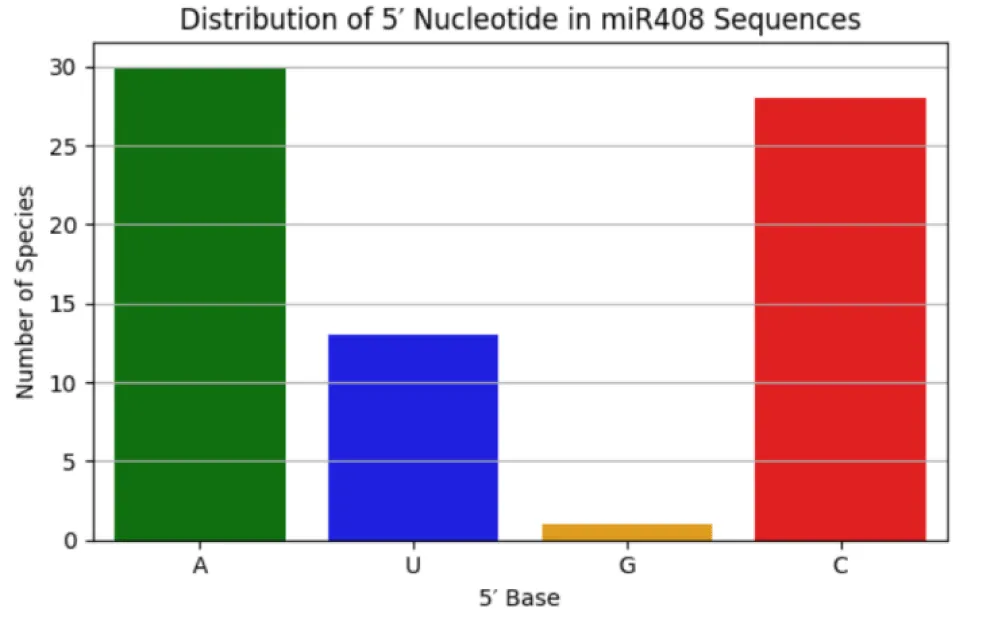
Since the 5′ nucleotide strongly influences AGO loading, the distribution of nucleotides at this position was calculated using a custom Python script in Google Colab. Bar plots were generated to illustrate the frequency of 5′-end bases across mature miR408 sequences.
Phylogenetic analyses
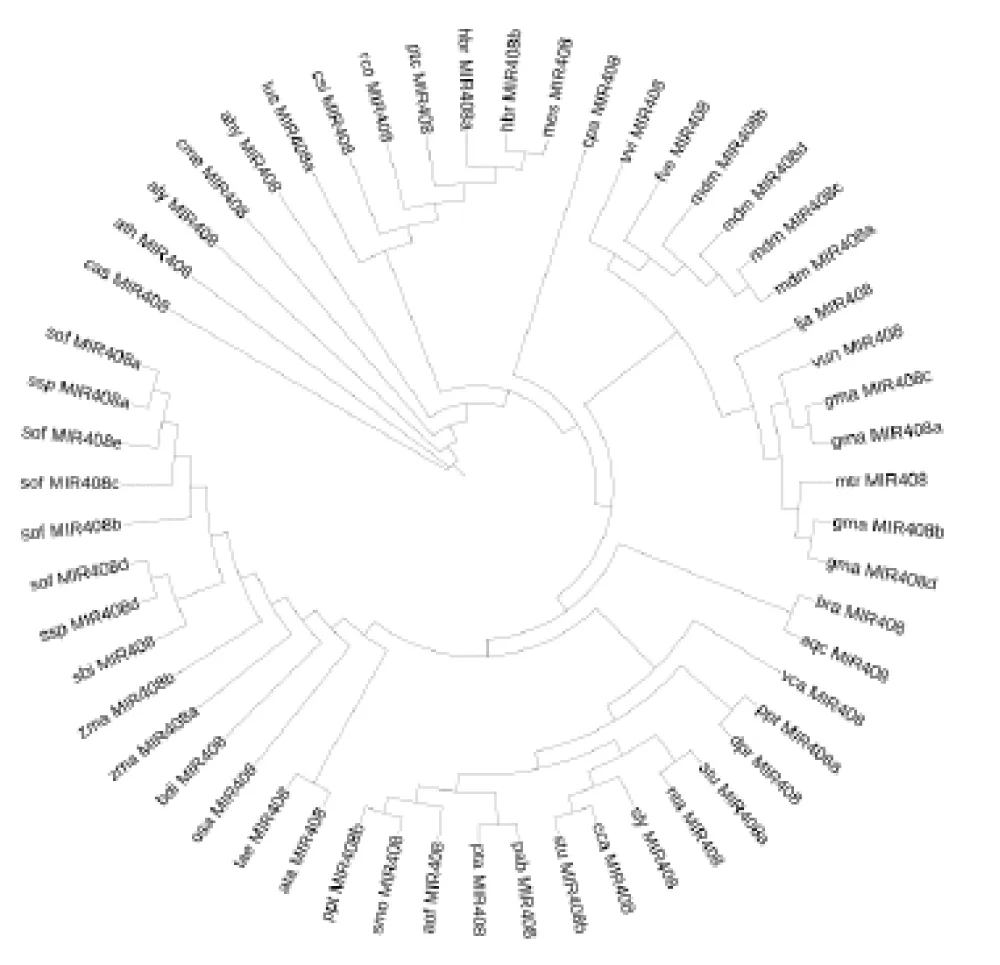
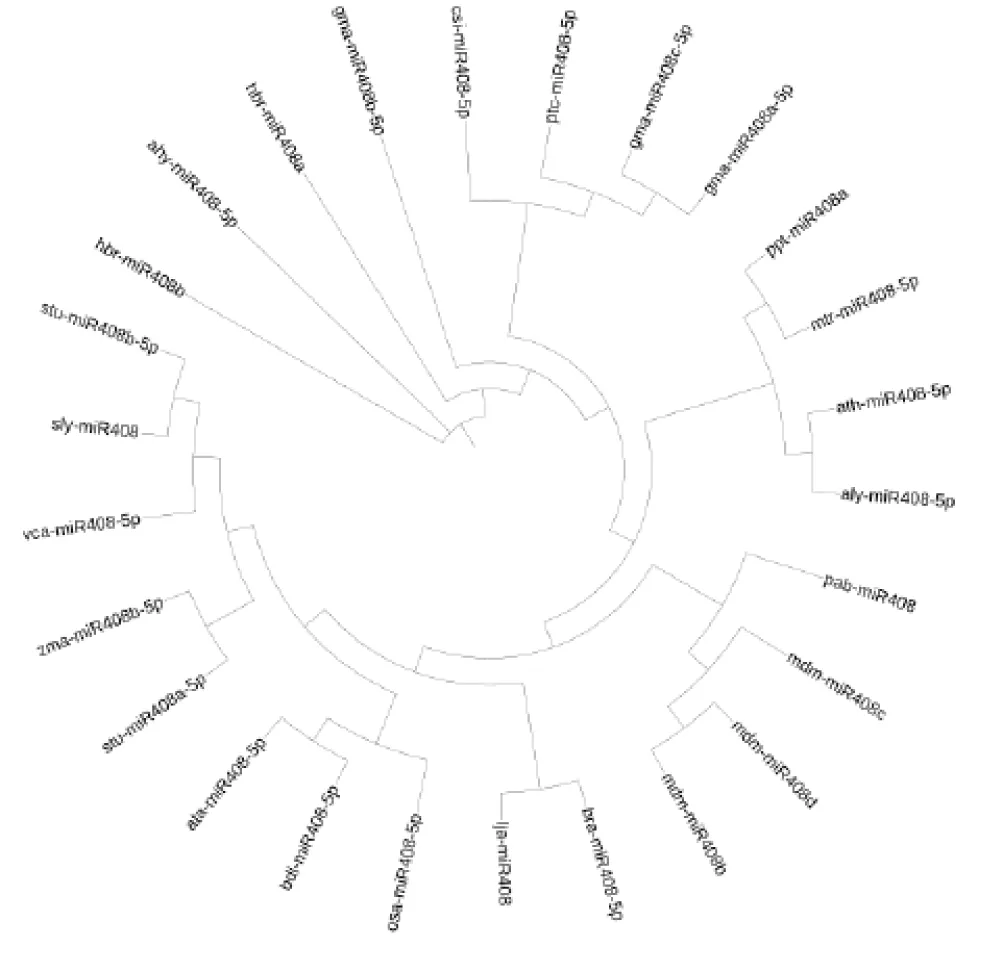
Precursor miRNA phylogeny: The 55 precursor miR408 sequences were aligned using MAFFT (https://mafft.cbrc.jp/alignment/server/index.html) [43]. Maximum likelihood phylogenies were reconstructed with the IQ-TREE web server (http://iqtree.cibiv.univie.ac.at/) [44,45], employing ModelFinder [46] for model selection and 5000 ultrafast bootstrap replicates [47] for branch support. Resulting trees in Newick format were visualized in iTOL v7.2 (https://itol.embl.de/) [48].
Mature miRNA phylogeny: Mature miR408-3p and 5p sequences were separately aligned in MAFFT, and phylogenetic trees were generated using IQ-TREE with identical parameters as above. Visualization was again performed in iTOL v7.2.
Precursor structural analysis
Secondary structures of precursor miR408 sequences were predicted using the RNAfold web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) [31,33]. Minimum free energy (MFE) values were recorded, while GC content and precursor length were calculated using a custom Python script in Google Colab. Scatterplots and bar plots were generated to evaluate trends in sequence stability.
Prediction of human targets
Potential cross-kingdom targets of plant miR408 were identified using psRNATarget (2017 update) (https://www.zhaolab.org/psRNATarget/) [49]. The consensus sequence was queried against the Homo sapiens cDNA library. Predicted targets were filtered for cleavage inhibition interactions, and the resulting dataset is provided in Supplementary Table 2.
Target site localization in human transcripts
The precise genomic coordinates and transcript regions (CDS, 5′ UTR, 3′ UTR) of the five high-confidence human target genes (AGO1, ZNF74, MAP6D1, SEMA5A, and ONECUT2) were retrieved from the Ensembl Genome Browser (https://www.ensembl.org) using MANE Select canonical transcripts. For each gene, the mature miR408 sequence was aligned against the full cDNA to identify the exact binding location. The genomic coordinates were validated against the GRCh38 human genome assembly. Based on this analysis, the target regions were classified as coding sequence (CDS), 5′ untranslated region (5′ UTR), or 3′ untranslated region (3′ UTR).
Target site alignment validation
To validate predicted interactions, pairwise alignments were conducted between miR408 and human target sites using a custom Python script in Google Colab. Seed region complementarity and 3′ compensatory pairing were specifically assessed, with Watson–Crick matches, wobble pairings, and mismatches recorded [27,38,40].
Gene function annotation
Functional information for predicted human targets was obtained from the Human Protein Atlas (version 24) (https://www.proteinatlas.org/) [51]. Genes were categorized into biological and molecular functions, enabling assessment of their regulatory relevance.
Functional enrichment analysis
To assess the broader significance of predicted targets, functional enrichment was first attempted using ShinyGO v0.76 (http://bioinformatics.sdstate.edu/go/) [52]. However, due to limited enrichment terms, further analysis was performed with the DAVID Functional Annotation Tool v6.8 (https://david.ncifcrf.gov/) [53,54]. Gene Ontology (GO) categories for Biological Process (BP) and Cellular Component (CC), along with UniProt Keywords, were retrieved. Multiple testing corrections (Bonferroni, FDR, Benjamini) were applied, and significantly enriched terms were reported.
Chromosomal mapping of human targets
The genomic locations of the predicted human target genes of miR408-3p were retrieved from publicly available databases. Gene coordinates, cytogenetic band positions, exon–intron structures, and strand orientation were collected from the NCBI Gene Database (https://www.ncbi.nlm.nih.gov/gene/) and the Ensembl Genome Browser (https://www.ensembl.org/), using the GRCh38/hg38 reference assembly. Chromosomal positions were recorded for each high-confidence target gene (AGO1, ZNF74, MAP6D1, SEMA5A, ONECUT2), and representative loci were used to illustrate their distribution across the human genome.
Results
Retrieval of miR408 sequences
From the miRBase database (v22), a total of 72 mature and 55 precursor miR408 sequences were retrieved (Table 2). Among the mature sequences, 17 were annotated directly as 3p and 5p arms, while the remaining 38 required assignment based on their annotated hairpin structures. This classification revealed 46 sequences belonging to the 3p arm and 26 sequences to the 5p arm (Table 3). These datasets formed the basis for further taxonomic, structural, and functional analyses.
Taxonomic distribution of miR408-encoding species
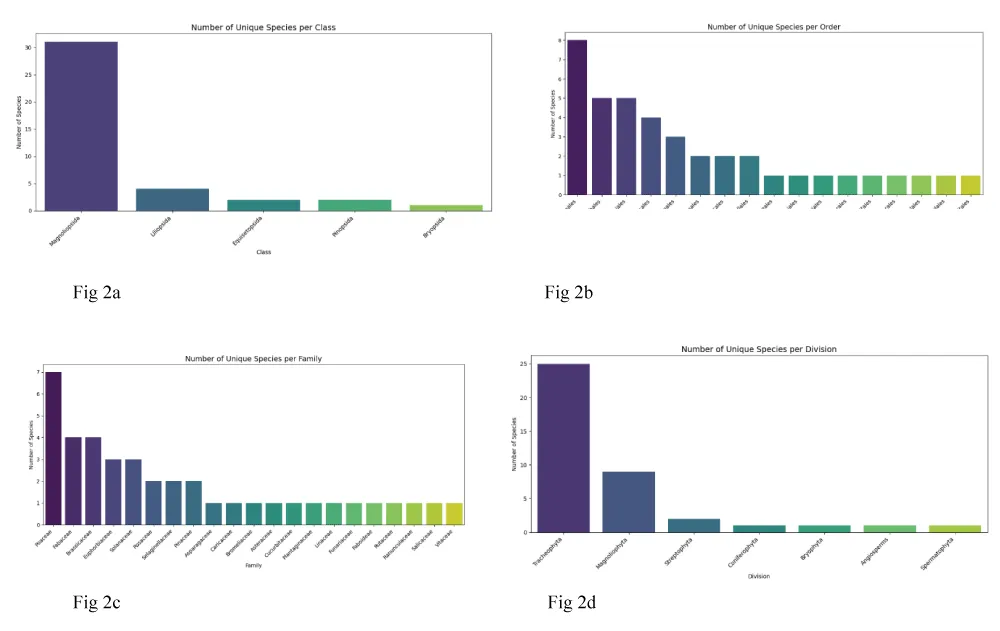
miR408 was identified across 40 plant species, spanning diverse taxonomic groups. The highest species representation was observed in the family Poaceae (7 species), order Poales (8 species), class Magnoliopsida (31 species), and division Tracheophyta (25 species) (Figure 2a-d; Supplementary Table 1). This indicates that miR408 is widely conserved across flowering plants, with strong representation in monocots such as cereals, highlighting its potential importance in fundamental plant processes.
Positional nucleotide conservation
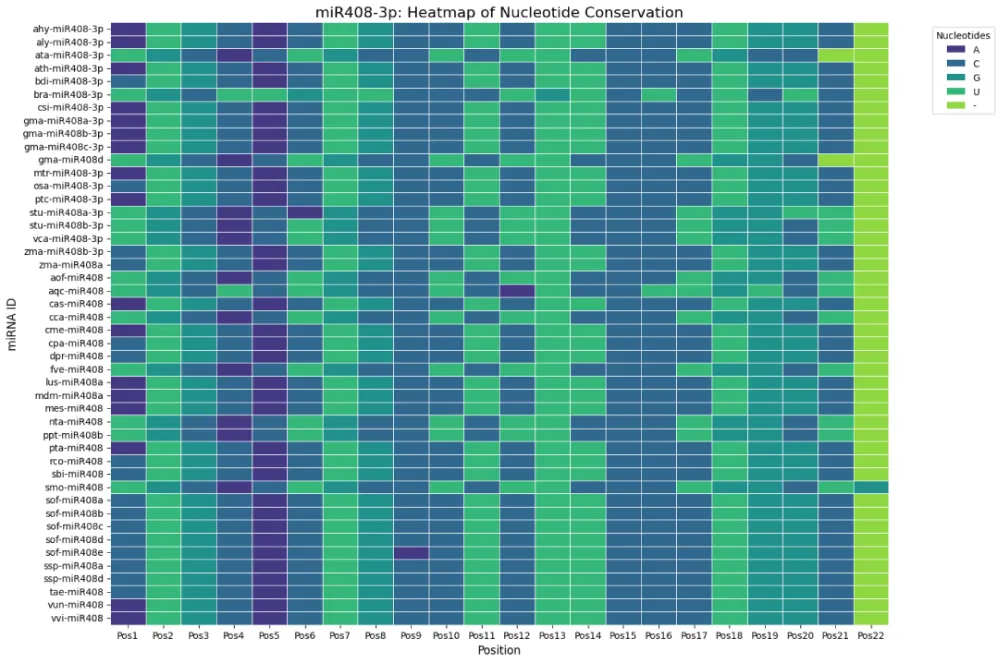
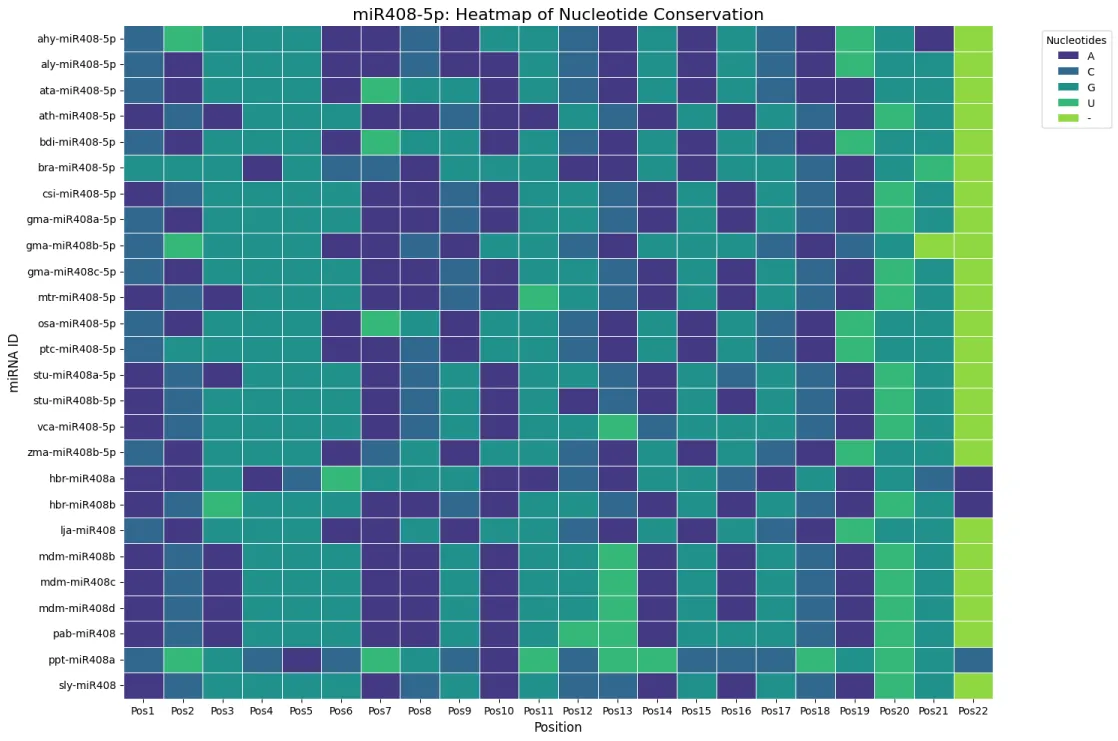
Heatmap analyses of nucleotide distributions revealed higher sequence conservation in the 3p arm compared to the 5p arm. Complete conservation was observed at position 15 (cytosine) in all 3p sequences, with additional conservation at positions 9 (mostly cytosine) and 13 (predominantly uridine) (Figure 3a). By contrast, the 5p arm showed greater variability across positions, consistent with relaxed evolutionary constraints (Figure 3b). Sequence logos generated by WebLogo further confirmed these observations, providing a clear visual representation of conserved motifs (Figure 3c,d). These findings suggest that the 3p strand plays a more conserved functional role, while the 5p strand may have undergone lineage-specific diversification.
AGO compatibility prediction
Analysis of the 5′ terminal nucleotide distribution provided insights into AGO loading preferences. The majority of 3p sequences began with uridine (n = 21), suggesting preferential binding to AGO1 and AGO10. In contrast, 5p sequences were enriched for adenosine (n = 11) and cytidine (n = 10), corresponding to potential associations with AGO2–AGO9 and AGO5, respectively (Figure 4; Tables 4,5). This divergence between 3p and 5p arms highlights possible functional specialization, with the conserved 3p strand showing canonical AGO1/AGO10 loading, and the variable 5p strand potentially engaging with multiple AGO pathways.
Phylogenetic analysis
Phylogenetic trees constructed from precursor and mature miRNA408 sequences revealed distinct evolutionary patterns. The precursor tree, built from 55 sequences, showed strong family-level clustering with well-supported monophyletic clades for Poaceae, Brassicaceae, and Fabaceae (Figure 5). Basal taxa such as Selaginella moellendorffii and Pinus taeda were positioned as outgroups, consistent with their early divergence.
In contrast, mature sequence phylogenies revealed higher conservation in 3p strands than in 5p strands. The 3p alignment contained 59% invariant sites, producing tight clusters with strong family-level resolution (Figure 6). The 5p alignment, however, showed only 55% invariant sites with greater variability, resulting in weaker clade resolution and mixed lineage clustering (Figure 7). Together, these results reinforce the deep evolutionary conservation of pre-miR408 and mature 3p strands, while supporting functional diversification in the 5p arm.
Precursor structural features
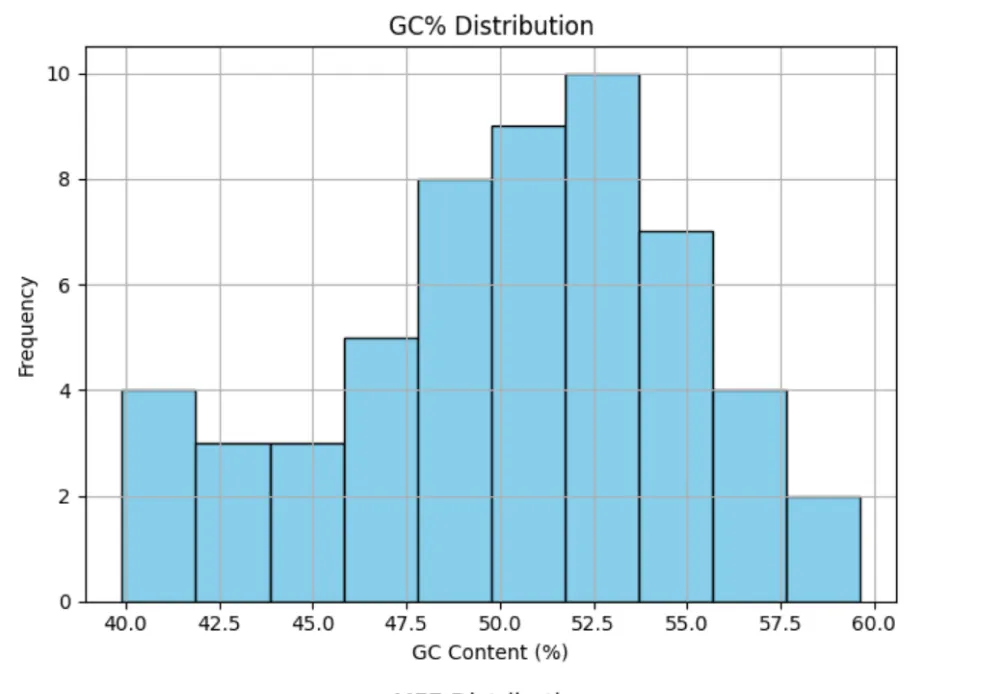
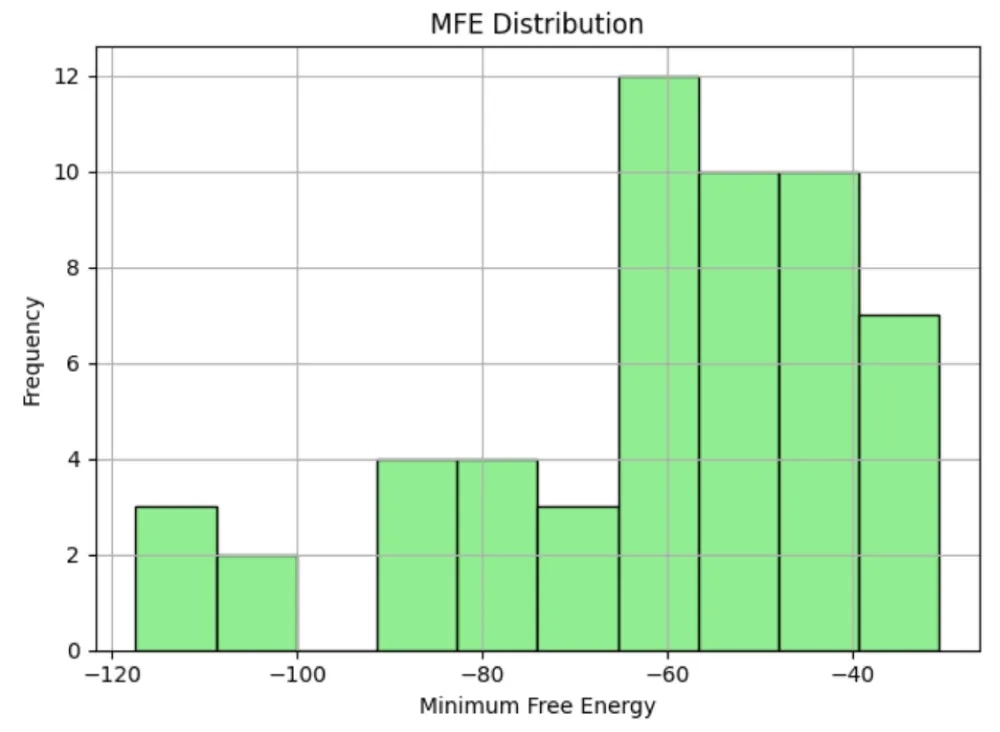
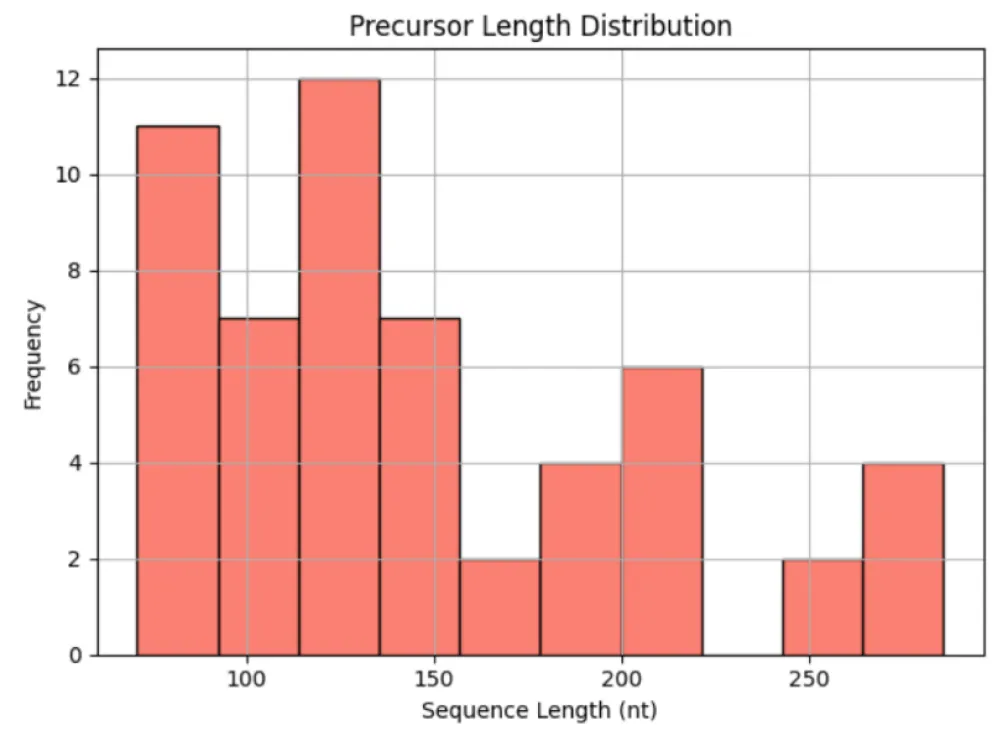
Structural analysis of 55 precursor miR408 sequences revealed considerable variability in length (71–286 nt), GC content (39.91% - 59.62%), and minimum free energy (MFE; −117.4 to −30.6 kcal/mol) (Table 6). Saccharum officinarum showed the most stable precursors (MFE −117.4 kcal/mol), whereas Solanum tuberosum had the least stable (−30.6 kcal/mol). Trends indicated that higher GC content correlated with greater structural stability (Figure 8a-c). These findings suggest that precursor stability, while variable, may play an important role in the regulation and efficiency of miR408 processing across different taxa.
Human target prediction
The consensus mature sequence of miR408-3p (5′-AUGAACUGCCUCUUCCCGGCG-3′) was used for cross-kingdom target prediction in Homo sapiens. psRNATarget analysis identified 68 potential human targets, of which 58 were categorized as cleavage-inhibition interactions (Supplementary Table 3). Alignment analyses revealed five high-confidence targets with strong seed and 3′ compensatory binding: AGO1, ZNF74, MAP6D1, SEMA5A, and ONECUT2 (Table 7). These genes are functionally linked to RNA silencing (AGO1), transcriptional regulation (ZNF74, ONECUT2), cytoskeletal organization (MAP6D1), and neuronal signaling (SEMA5A). The diversity of these predicted interactions suggests a plausible role for dietary miR408 in modulating mammalian gene expression.
Chromosomal mapping of human targets: Chromosomal mapping revealed that the predicted human targets of miR408-3p are distributed across multiple chromosomes, indicating no genomic clustering but rather a dispersed pattern of target localization. AGO1 is located on chromosome 1 at cytogenetic band 1p34.3, ZNF74 on chromosome 22 at 22q11.21, and SEMA5A at 5p15.31. ZNF74 lies in a region often associated with neurodevelopmental phenotypes, while SEMA5A is positioned in a region linked to developmental disorders. The remaining targets, MAP6D1 and ONECUT2, are located on chromosomes 3 and 18, respectively. This widespread genomic distribution suggests that miR408-3p may modulate diverse biological pathways across the human genome rather than acting via genomic clustering. The detailed chromosomal locations, cytogenetic bands, genomic coordinates, and exon counts of these high-confidence targets are summarized in Table 8.
Target site mapping in human transcripts
To gain deeper insight into the potential regulatory mechanisms of miR408 on human targets, we performed a detailed analysis of the exact binding sites on the canonical transcripts of the five high-confidence target genes (AGO1, ZNF74, MAP6D1, SEMA5A, and ONECUT2). The MANE Select transcript for each gene was retrieved from the Ensembl Genome Browser, and the mature miR408 sequence was aligned to the full cDNA sequence to identify precise binding locations. The genomic coordinates were validated using the GRCh38 human reference assembly. Our analysis revealed that the miR408 target sites are distributed across different transcript regions: AGO1 (ENST00000373204.6) and SEMA5A (ENST00000382496.10) exhibited binding sites within the coding sequence (CDS), indicating a potential for direct interference with protein translation or mRNA stability. ZNF74 (ENST00000400451.7) was found to be targeted in the 5′ untranslated region (5′ UTR), which may affect translation initiation. Meanwhile, MAP6D1 (ENST00000318631.8) and ONECUT2 (ENST00000491143.3) displayed target sites in the 3′ untranslated region (3′ UTR), suggesting a role in mRNA stability and post-transcriptional regulation. These findings are summarized in Table 9 and provide further computational support for the hypothesis that miR408 may exert cross-kingdom gene regulation through diverse mechanisms.
Functional annotation and enrichment
Functional annotation of predicted targets, performed using DAVID and the Human Protein Atlas, indicated that miR408-3p targets span a broad spectrum of cellular processes. Key functional categories included transcriptional regulation (ONECUT2, ZNF74, MEF2C), RNA interference and silencing (AGO1), neural development (SEMA5A, MAP6D1), and immune regulation (TREML2, SKAP1, MAVS). Although enrichment analysis did not yield statistically significant pathways, the convergence of targets across transcriptional, neuronal, and immune functions supports the hypothesis that miR408 could influence multiple physiological systems if taken up in mammalian cells (Supplementary Tables 5-7).
Discussion
MicroRNA408 (miR408) is one of the most conserved plant miRNAs, with established roles in development, vascular differentiation, redox balance, and abiotic stress responses. In this study, we conducted a comprehensive comparative analysis of miR408 across plant species, focusing on sequence conservation, AGO-binding preferences, phylogenetic relationships, structural stability, and potential cross-kingdom regulatory activity in humans.
Our retrieval of 72 mature and 55 precursor miR408 sequences from miRBase confirmed the broad taxonomic distribution of this miRNA family. The predominance of the 3p arm, observed both in annotation frequency and sequence conservation, is consistent with models of asymmetric arm selection during miRNA biogenesis. In particular, the invariant cytosine at position 15 of 3p sequences highlights a conserved structural feature likely critical for target recognition. By contrast, the higher variability in 5p sequences suggests lineage-specific or condition-specific adaptations, reflecting relaxed selection pressures relative to the conserved 3p strand.
AGO-binding preferences further emphasized this asymmetry. Most 3p sequences began with uridine, a canonical signal for AGO1/AGO10 loading, whereas 5p sequences frequently started with adenosine or cytidine, corresponding to alternative AGO associations. These patterns reinforce the dominance of the 3p strand in canonical silencing pathways while suggesting that the 5p strand may serve auxiliary or context-specific functions in certain species.
Phylogenetic analyses revealed strong clustering of precursor sequences into family-level clades, particularly within Poaceae, Fabaceae, and Brassicaceae. Mature sequence trees further highlighted the stability of the 3p arm, which showed tight family-specific groupings and high conservation across angiosperms. In contrast, the weaker clustering of 5p sequences supports their evolutionary diversification. Together, these results suggest that miR408-3p has been maintained as the functional strand under strong selective pressure, while 5p diversification may allow for adaptive regulatory flexibility.
Structural analysis of precursors revealed wide variability in length, GC content, and minimum free energy. Stable precursors with high GC% were more frequent in cereals such as Saccharum officinarum and Triticum aestivum, while dicot species like Solanum tuberosum showed shorter, less stable precursors. These findings suggest that structural stability of pre-miR408 may influence processing efficiency and abundance, and could contribute to observed differences in strand bias across taxa. Transcript per million (TPM) values have been reported for miR408 amid wide variation across plant species, ranging from ~0.8 TPM in inactive sugarcane buds to ~9.1 TPM in active buds (Saccharum officinarum) [76], and up to 147 TPM in Capsicum eximium flowers and small fruits (Capsicum spp.) [77], reflecting moderate abundance and mobility of miR408 transcripts in developing tissues.
Extending the analysis to humans, psRNATarget predictions identified 68 possible miR408-3p targets, with five high-confidence candidates: AGO1, ZNF74, MAP6D1, SEMA5A, and ONECUT2. These genes are functionally diverse, spanning RNA silencing, transcriptional regulation, cytoskeletal stability, and neuronal signaling. The identification of AGO1 is particularly intriguing, as it suggests a potential regulatory feedback loop whereby a plant miRNA might modulate the very machinery involved in small RNA function. While computational predictions require experimental validation, the convergence of targets in regulatory and neuronal pathways supports a plausible cross-kingdom influence.
A further layer of analysis mapped the exact miR408 binding sites within the transcript structure of each high-confidence target. AGO1 and SEMA5A were found to be targeted within their coding sequences (CDS), which may allow for direct translational repression or mRNA destabilization. ZNF74 exhibited a binding site in the 5′ UTR, potentially affecting translation initiation efficiency, whereas MAP6D1 and ONECUT2 were targeted in the 3′ UTR, consistent with classical post-transcriptional regulation mechanisms. The diversity in target site location suggests that miR408 may exert cross-kingdom regulation through multiple mechanistic pathways (Table 8). These results strengthen the hypothesis that miR408 could modulate mammalian gene expression in a complex, multi-faceted manner.
Chromosomal mapping further revealed that miR408-3p targets are distributed across multiple chromosomes, including regions implicated in neurodevelopmental disorders, such as SEMA5A at 5p15.3 and ZNF74 at 22q11. The former has been associated with autism spectrum disorder and intellectual disability, while the latter is linked to DiGeorge syndrome. This spatial distribution supports the potential relevance of miR408 targets in neurological pathways and broad cellular processes.
Functional annotation of these targets revealed enrichment in biological themes such as transcriptional regulation, epigenetic control, immune signaling, and synaptic function. Although no Gene Ontology (GO) terms reached statistical significance, the convergence of functional categories across independent targets suggests that miR408-3p could, if systemically absorbed, exert meaningful influence on mammalian gene networks. Supporting this, recent evidence from Camellia japonica showed that extracellular vesicle–packaged miR408 enhances fibroblast proliferation and wound healing, demonstrating the potential functional activity of plant-derived miRNAs in humans.
Finally, emerging studies on miRNA-encoded peptides (miPEPs), such as miPEP408, provide an additional regulatory layer. In Arabidopsis thaliana, miPEP408 enhances expression of its cognate miRNA, forming a self-regulatory feedback loop at the level of pri-miRNA transcripts. This dual functionality RNA-based silencing and peptide-based regulation,underscores the evolutionary significance of miR408 and highlights the need for integrated approaches to fully understand its biological roles.
Taken together, our findings establish miR408-3p as the dominant functional strand, characterized by evolutionary conservation, canonical AGO loading, structural stability, strong target complementarity, and diverse modes of action in putative human targets. Future work should focus on experimental validation of miR408 stability and systemic uptake in mammalian systems, its tissue distribution, and its functional regulatory impact. Integrating computational, molecular, and translational approaches will illuminate both the evolution of plant miRNAs and their intriguing cross-kingdom functions.
Conclusion
This study provides a comprehensive computational analysis of miR408 across diverse plant species, revealing its remarkable evolutionary conservation, arm-specific biases, and intriguing potential for cross-kingdom interactions. Our findings consistently highlight the dominance of the 3p arm, which exhibited strong nucleotide conservation, canonical AGO1/AGO10 loading preferences, and tighter phylogenetic clustering compared to the more variable 5p arm. Structural analyses of precursor sequences further underscored the importance of hairpin length, GC content, and folding stability in influencing miRNA408 biogenesis and strand selection across taxa.
Extending beyond plants, cross-kingdom target predictions identified five high-confidence human gene targets, AGO1, ZNF74, MAP6D1, SEMA5A, and ONECUT2, implicating miR408-3p in the potential modulation of regulatory, neuronal, and cytoskeletal pathways within mammalian systems. A novel layer of analysis mapped these targets to distinct transcript regions: AGO1 and SEMA5A in the coding sequence (CDS), ZNF74 in the 5′ untranslated region (5′ UTR), and MAP6D1 and ONECUT2 in the 3′ UTR. This suggests multiple mechanisms through which miR408-3p could regulate gene expression, such as translational repression, interference with translation initiation, or modulation of mRNA stability. Although functional enrichment analysis did not reach statistical significance, the convergence of target gene functions in biologically coherent pathways lends further support to the hypothesis of cross-kingdom regulatory potential.
Moreover, recent studies on miRNA-encoded peptides (miPEPs), such as miPEP408, expand the regulatory dimension of miR408 beyond RNA silencing to include peptide-mediated feedback mechanisms. These dual functional roles highlight the evolutionary complexity and potential adaptability of miR408 as a regulatory molecule.
Overall, our results position miR408-3p as a valuable model for studying plant miRNA conservation, biogenesis, and cross-kingdom regulatory functions. However, translating these computational predictions into biological relevance requires rigorous experimental validation. Future research should focus on verifying the stability and systemic uptake of plant miR408 in mammalian systems, determining its tissue-specific distribution, and experimentally assessing its impact on the expression of target genes. Such integrated molecular and translational studies will be essential to fully elucidate the role of plant-derived miRNAs in inter-kingdom communication and their potential applications in therapeutics and human health.
Availability of data and materials
The data supporting the findings of this study are available from miRBase v22, psRNATarget, and public repositories cited within the manuscript.
SUPPLEMENTARY INFORMATION
Supplementary_Table_1 - mir408_mature&precursor_info_miRBase
Supplementary_Table_2 - psRNATarget_consensus_3p_targets
Supplementary_Table_3 - miRNA_detailed_alignment_analysis
Supplementary_Table_4 - top5_miRNA_targets_with_alignment
Supplementary_Table_5 - 3p_target_geneInfo_ShinyGO
Supplementary_Table_6 - DAVID
Supplementary_Table_7 - 3p_target_HPA_details
- Gao Y, Feng B, Gao C, Zhang H, Wen F, Tao L, et al. The evolution and functional roles of miR408 and its targets in plants. Int J Mol Sci. 2022;23(1):530. Available from: https://doi.org/10.3390/ijms23010530
- Qin L, Xu P, Jiao Y. Evolution of plant conserved microRNAs after whole-genome duplications. Genome Biol Evol. 2025;17(3):evaf045. Available from: https://doi.org/10.1093/gbe/evaf045
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. Available from: https://doi.org/10.1016/j.cell.2009.01.002
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136(4):669–87. Available from: https://doi.org/10.1016/j.cell.2009.01.046
- Axtell MJ. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol. 2013;64(1):137–59. Available from: https://doi.org/10.1146/annurev-arplant-050312-120043
- Dai X, Zhuang Z, Zhao PX. Computational analysis of miRNA targets in plants: current status and challenges. Brief Bioinform. 2011;12(2):115–21. Available from: https://doi.org/10.1093/bib/bbq065
- Lewis BP, Shih I-H, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–98. Available from: https://doi.org/10.1016/s0092-8674(03)01018-3
- Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14(8):475–88. Available from: https://doi.org/10.1038/nrm3611
- Solanki M, Sinha A, Shukla LI. The miR408 expression in scutellum derived somatic embryos of Oryza sativa L. ssp. indica varieties: media and regenerating embryos. Plant Cell Tissue Organ Cult PCTOC. 2019;138(1):53–66. Available from: https://doi.org/10.1007/s11240-019-01602-w
- Sunkar R, Li YF, Jagadeeswaran G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012;17(4):196–203. Available from: https://doi.org/10.1016/j.tplants.2012.01.010
- Rogers K, Chen X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell. 2013;25(7):2383–99. Available from: https://doi.org/10.1105/tpc.113.113159
- Axtell MJ, Meyers BC. Revisiting criteria for plant microRNA annotation in the era of big data. Plant Cell. 2018;30(2):272–84. Available from: https://doi.org/10.1105/tpc.17.00851
- Chávez Montes RA, Rosas-Cárdenas DFF, De Paoli E, Accerbi M, Rymarquis LA, Mahalingam G, et al. Sample sequencing of vascular plants demonstrates widespread conservation and divergence of microRNAs. Nat Commun. 2014;5(1):3722. Available from: https://doi.org/10.1038/ncomms4722
- Song Z, Zhang L, Wang Y, Li H, Li S, Zhao H, et al. Constitutive expression of miR408 improves biomass and seed yield in Arabidopsis. Front Plant Sci. 2018;8:2114. Available from: https://doi.org/10.3389/fpls.2017.02114
- Sunkar R, Kapoor A, Zhu JK. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006;18(8):2051–65. Available from: https://doi.org/10.1105/tpc.106.041673
- Abdel-Ghany SE, Pilon M. MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J Biol Chem. 2008;283(23):15932–45. Available from: https://doi.org/10.1074/jbc.m801406200
- Guo Y, Wang S, Yu K, Wang HL, Xu H, Song C, et al. Manipulating microRNA miR408 enhances both biomass yield and saccharification efficiency in poplar. Nat Commun. 2023;14(1):4285. Available from: https://doi.org/10.1038/s41467-023-39930-3
- Abdel-Ghany SE, Müller-Moulé P, Niyogi KK, Pilon M, Shikanai T. Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell. 2005;17(4):1233–51. Available from: https://doi.org/10.1105/tpc.104.030452
- Zhao X, Hong PD, Wu JY, Chen XB, Ye XG, Pan YY, et al. The tae-miR408-mediated control of TaTOC1 gene transcription is required for the regulation of heading time in wheat. Plant Physiol. 2016;170(3):1578–94. Available from: https://doi.org/10.1104/pp.15.01216
- Xu X, Zhang C, Xu X, Cai R, Guan Q, Chen X, et al. Riboflavin mediates m6A modification targeted by miR408, promoting early somatic embryogenesis in longan. Plant Physiol. 2023;192(3):1799–820. Available from: https://doi.org/10.1093/plphys/kiad139
- Rong F, Lv Y, Deng P, Wu X, Zhang Y, Yue E, et al. Switching action modes of miR408-5p mediates auxin signaling in rice. Nat Commun. 2024;15(1):2525. Available from: https://doi.org/10.1038/s41467-024-46765-z
- Pan J, Huang D, Guo Z, Kuang Z, Zhang H, Xie X, et al. Overexpression of microRNA408 enhances photosynthesis, growth, and seed yield in diverse plants. J Integr Plant Biol. 2018;60(4):323–40. Available from: https://doi.org/10.1111/jipb.12634
- Kuo YW, Lin JS, Li YC, Jhu MY, King YC, Jeng ST. MicroR408 regulates defense response upon wounding in sweet potato. J Exp Bot. 2019;70(2):469–83. Available from: https://doi.org/10.1093/jxb/ery381
- Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138(4):738–49. Available from: https://doi.org/10.1016/j.cell.2009.06.014
- Zhang H, Li Y, Zhu JK. Developing naturally stress-resistant crops for a sustainable agriculture. Nat Plants. 2018;4(12):989–96. Available from: https://doi.org/10.1038/s41477-018-0309-4
- Ma C, Burd S, Lers A. miR408 is involved in abiotic stress responses in Arabidopsis. Plant J. 2015;84(1):169–87. Available from: https://doi.org/10.1111/tpj.12999
- Ebrahimi Khaksefidi R, Mirlohi S, Khalaji F, Fakhari Z, Shiran B, Fallahi H, et al. Differential expression of seven conserved microRNAs in response to abiotic stress and their regulatory network in Helianthus annuus. Front Plant Sci. 2015;6:741.Available from: https://doi.org/10.3389/fpls.2015.00741
- Kumar RS, Sinha H, Datta T, Asif MH, Trivedi PK. microRNA408 and its encoded peptide regulate sulfur assimilation and arsenic stress response in Arabidopsis. Plant Physiol. 2023;192(2):837–56. Available from: https://doi.org/10.1093/plphys/kiad033
- Kumar RS, Datta T, Sinha H, Trivedi PK. Light-dependent expression and accumulation of miR408-encoded peptide, miPEP408, is regulated by HY5 in Arabidopsis. Biochem Biophys Res Commun. 2024;706:149764. Available from: https://doi.org/10.1016/j.bbrc.2024.149764
- Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6(1):26. Available from: https://doi.org/10.1186/1748-7188-6-26
- Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155–62. Available from: https://doi.org/10.1093/nar/gky1141
- Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36(Web Server issue):W70-4. Available from: https://doi.org/10.1093/nar/gkn188
- Arazi T, Khedia J. Tomato microRNAs and their functions. Int J Mol Sci. 2022;23(19):11979. Available from: https://doi.org/10.3390/ijms231911979
- Zhang L, Hou D, Chen X, Li D, Zhu L, Zhang Y, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012;22(1):107–26. Available from: https://doi.org/10.1038/cr.2011.158
- Samad AFA, Kamaroddin MF, Sajad M. Cross-kingdom regulation by plant microRNAs provides novel insight into gene regulation. Adv Nutr. 2021;12(1):197–211. Available from: https://doi.org/10.1093/advances/nmaa095
- Zhou Z, Li X, Liu J, Dong L, Chen Q, Liu J, et al. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. 2015;25(1):39–49. Available from: https://doi.org/10.1038/cr.2014.130
- Chin AR, Fong MY, Somlo G, Wu J, Swiderski P, Wu X, et al. Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res. 2016;26(2):217–28. Available from: https://doi.org/10.1038/cr.2016.13
- Kosek DM, Petzold K, Andersson ER. Mapping effective microRNA pairing beyond the seed using abasic modifications. Nucleic Acids Res. 2025;53(8):gkaf364. Available from: https://doi.org/10.1093/nar/gkaf364
- Witwer KW. Alternative miRNAs? Human sequences misidentified as plant miRNAs in plant studies and in human plasma. F1000Research. 2018;7:244. Available from: https://doi.org/10.12688/f1000research.14060.1
- Liao P, Li S, Cui X, Zheng Y. A comprehensive review of web-based resources of non-coding RNAs for plant science research. Int J Biol Sci. 2018;14(8):819–32. Available from: https://doi.org/10.7150/ijbs.24593
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188-90. Available from: https://doi.org/10.1101/gr.849004
- Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20(4):1160–6. Available from: https://doi.org/10.1093/bib/bbx108
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32(1):268–74. Available from: https://doi.org/10.1093/molbev/msu300
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44(W1):W232–5. Available from: https://doi.org/10.1093/nar/gkw256
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14(6):587–9. Available from: https://doi.org/10.1038/nmeth.4285
- Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35(2):518–22. Available from: https://doi.org/10.1093/molbev/msx281
- Letunic I, Bork P. Interactive Tree of Life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024;52(W1):W78–82. Available from: https://doi.org/10.1093/nar/gkae268
- Dai X, Zhuang Z, Zhao PX. psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018;46(W1):W49–54. Available from: https://doi.org/10.1093/nar/gky316
- Hall MH, Wang PY, Pham TM, Bartel DP. Functional microRNA targeting without seed pairing [Internet]. Molecular Biology; 2025 [cited 2025 Jun 6]. Available from: https://doi.org/10.1101/2025.03.03.641270
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. A human tissue proteomic map. Nat Methods. 2015;12(4):288–288. Available from: https://doi.org/10.1126/science.1260419
- Ge SX, Jung D, Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Valencia A, editor. Bioinformatics. 2020;36(8):2628–9. Available from: https://doi.org/10.1093/bioinformatics/btz931
- Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022;50(W1):W216–21. Available from: https://doi.org/10.1093/nar/gkac194
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. Available from: https://doi.org/10.1038/nprot.2008.211
- Mosca-Boidron AL, Gueneau L, Huguet G, Goldenberg A, Henry C, Gigot N, et al. A de novo microdeletion of SEMA5A in a boy with autism spectrum disorder and intellectual disability. Eur J Hum Genet. 2016 Jun;24(6):838–43. Available from: https://doi.org/10.1038/ejhg.2015.211
- Liu C, Li A, Guo Z, Chen N, Wang Y, Tang W, et al. MicroRNA analysis reveals two modules that antagonistically regulate xylem tracheary element development in Arabidopsis. Plant Cell. 2024;37(1):koaf011. Available from: https://doi.org/10.1093/plcell/koaf011
- Yu Y, Jia T, Chen X. The “how” and “where” of plant microRNAs. New Phytol. 2017;216(4):1002–17. Available from: https://doi.org/10.1111/nph.14834
- Bologna NG, Voinnet O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev Plant Biol. 2014;65(1):473–503. Available from: https://doi.org/10.1146/annurev-arplant-050213-035728
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, et al. Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133(1):116–27. Available from: https://doi.org/10.1016/j.cell.2008.02.034
- Zhang Z, Liu X, Guo X, Wang XJ, Zhang X. Arabidopsis AGO3 predominantly recruits 24-nt small RNAs to regulate epigenetic silencing. Nat Plants. 2016;2(5):16049. Available from: https://doi.org/10.1038/nplants.2016.49
- Endo Y, Iwakawa H, Tomari Y. Arabidopsis ARGONAUTE7 selects miR390 through multiple checkpoints during RISC assembly. EMBO Rep. 2013;14(7):652–8. Available from: https://doi.org/10.1038/embor.2013.73
- Thieme CJ, Schudoma C, May P, Walther D. Give it AGO: the search for miRNA-Argonaute sorting signals in Arabidopsis thaliana indicates a relevance of sequence positions other than the 5′-position alone. Front Plant Sci [Internet]. 2012 [cited 2025 Jun 13];3. Available from: https://doi.org/10.3389/fpls.2012.00272
- Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505(7485):696–700. Available from: https://doi.org/10.1038/nature12756
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–15. Available from: https://doi.org/10.1093/nar/gkg595
- Shuaib M, Parsi KM, Thimma M, Adroub SA, Kawaji H, Seridi L, et al. Nuclear AGO1 regulates gene expression by affecting chromatin architecture in human cells. Cell Syst. 2019;9(5):446–458.e6. Available from: https://doi.org/10.1016/j.cels.2019.09.005
- Qian C, Yang Q, Rotinen M, Huang R, Kim H, Gallent B, et al. ONECUT2 acts as a lineage plasticity driver in adenocarcinoma as well as neuroendocrine variants of prostate cancer. Nucleic Acids Res. 2024;52(13):7740–60. Available from: https://doi.org/10.1093/nar/gkae547
- Grondin B, Bazinet M, Aubry M. The KRAB zinc finger gene encodes an RNA-binding protein tightly associated with the nuclear matrix. J Biol Chem. 1996;271(26):15458–67. Available from: https://doi.org/10.1074/jbc.271.26.15458
- Berto S, Perdomo-Sabogal A, Gerighausen D, Qin J, Nowick K. A consensus network of gene regulatory factors in the human frontal lobe. Front Genet [Internet]. 2016 [cited 2025 Jul 22];7. Available from: https://doi.org/10.3389/fgene.2016.00031
- Cuveillier C, Boulan B, Ravanello C, Denarier E, Deloulme JC, Gory-Fauré S, et al. Beyond neuronal microtubule stabilization: MAP6 and CRMPS, two converging stories. Front Mol Neurosci [Internet]. 2021. [cited 2025 Jul 22];14. Available from: https://doi.org/10.3389/fnmol.2021.665693
- Jin S, Kim JG, Kim HJ, Kim JY, Kim SH, Kang HC, et al. miRNA408 from Camellia japonica L. mediates cross-kingdom regulation in human skin recovery. Biomolecules. 2025;15(8):1108. Available from: https://doi.org/10.3390/biom15081108
- Lukasik A, Zielenkiewicz P. In silico identification of plant miRNAs in mammalian breast milk exosomes: a small step forward? Scaria V, editor. PLoS One. 2014;9(6):e99963. Available from: https://doi.org/10.1371/journal.pone.0099963
- Zhang L, Chen T, Yin Y, Zhang CY, Zhang YL. Dietary microRNA—a novel functional component of food. Adv Nutr. 2019;10(4):711–21. Available from: https://doi.org/10.1093/advances/nmy127
- Samad AFA, Kamaroddin MF, Sajad M. Cross-kingdom regulation by plant microRNAs provides novel insight into gene regulation. Adv Nutr. 2021;12(1):197–211. Available from: https://doi.org/10.1093/advances/nmaa095
- Liang G, Zhu Y, Sun B, Shao Y, Jing A, Wang J, et al. Assessing the survival of exogenous plant microRNA in mice. Food Sci Nutr. 2014;2(4):380–8. Available from: https://doi.org/10.1002/fsn3.113
- Couzigou JM, Lauressergues D, Bécard G, Combier JP. miRNA-encoded peptides (miPEPs): a new tool to analyze the roles of miRNAs in plant biology. RNA Biol. 2015;12(11):1178–80. Available from: https://doi.org/10.1080/15476286.2015.1094601
- Ormancey M, Thuleau P, Combier JP, Plaza S. The essentials on microRNA-encoded peptides from plants to animals. Biomolecules. 2023;13(2):206. Available from: https://doi.org/10.3390/biom13020206
- Ortiz-Morea FA, Vicentini R, Silva GF, Silva EM, Carrer H, Rodrigues AP, et al. Global analysis of the sugarcane microtranscriptome reveals a unique composition of small RNAs associated with axillary bud outgrowth. J Exp Bot. 2013;64(8):2307–20. Available from: https://doi.org/10.1093/jxb/ert089
- Lopez-Ortiz C, Peña-Garcia Y, Bhandari M, Abburi VL, Natarajan P, Stommel J, et al. Identification of miRNAs and their targets involved in flower and fruit development across domesticated and wild Capsicum species. Int J Mol Sci. 2021;22(9):4866. Available from: https://doi.org/10.3390/ijms22094866
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.



 Save to Mendeley
Save to Mendeley
